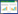中国科学院微生物研究所,中国微生物学会,中国菌物学会
文章信息
- 王飞, 李周坤, 周杰, 崔中利
- Fei Wang, Zhoukun Li, Jie Zhou, Zhongli Cui
- 定点突变对酰胺水解酶DamH可溶性表达和酶活的影响
- Effect of site-directed mutagenesis on soluble expression and specific activity of amide hydrolase DamH
- 微生物学报, 2015,55(12): 1584-1592
- Acta Microbiologica Sinica, 2015,55(12): 1584-1592
-
文章历史
- 收稿日期:2015-05-19
- 修回日期:2015-06-28
2 南京农业大学生命科学学院,农业部农业环境微生物工程重点实验室,江苏 南京 210095
2 Key Laboratory of Agricultural Environmental Microbiology, Ministry of Agriculture, Nanjing Agriculture University, Nanjing 210095, Jiangsu Province, China
氯代乙酰胺类除草剂因分子结构中具有酰胺基团(CONH-)而得名,因其高效、高选择性而在全球范围内广泛使用[1]。乙草胺[2’-乙基-6’-甲基-N-(乙氧基甲基)-2-氯代-N-乙酰苯胺]是氯代乙酰胺类除草剂的重要代表,主要用于一年生禾本科杂草和部分阔叶杂草的防除[2]。大量使用氯乙酰胺类除草剂导致的环境残留对水环境和农业生态系统构成不利影响[3],可导致小鼠形成肿瘤[4],造成对两栖动物的细胞毒性,染色体和DNA损伤[5],1994年被美国环保局(USEPA)列为B-2类致癌物[6, 7]。乙草胺的主要生物降解产物为CMEPA和2-甲基-6-乙基苯胺(MEA)[8]。侯颖等以小旱稗[Echinochloa crusgalli (L.)Beauv. var. austrojaponensis Ohwi]和马唐[Digitaria sanguinalis (L.)Scop.]两种杂草为试验靶标,对乙草胺及其代谢中间产物CMEPA和MEA的除草活性进行了测定,结果表明三种化合物对试验材料的抑制活性强弱程度依次为CMEPA>乙草胺>MEA,且低浓度的MEA(<20 mg/L)对两种杂草的生长似乎有刺激作用[9]。因此,CMEPA水解是消除环境中乙草胺残留毒性的重要步骤之一。
近年来,国内外研究者从不同的菌株中已克隆到一些催化CMEPA水解酶基因。Weiliang Shen等通过鸟枪法从Paracoccus sp. M-1中克隆到CMEPA水解酶PamH,该酶分子量大小为50 kDa,酶活在pH6.0-9.0之间稳定,最适反应温度为45 ℃[10]。Jun Zhang等从Paracoccus sp. FLN-7中克隆到一个新的CMEPA水解酶AmpA,该酶分子量49.6 kDa,最适反应温度40 ℃,最适反应pH7.5-8.0[11]。氨基酸序列分析表明这些酶都归属于酰胺酶(EC 3.4.11.2.),具有典型的酰胺酶保守序列GGSS和催化活性中心Lys-Ser-Ser。
本课题组前期从生产乙草胺农药厂活性污泥中分离到一株CMEPA高效水解菌株Delftia sp. T3-6[1],并成功克隆到催化该水解反应的酰胺水解酶DamH[12]。DamH分子量大小为32 kDa,最适底物为CMEPA,其Km和kcat值分别为0.197 mmol/L和2804.32 s-1。最适酶反应pH为6.5,最适酶反应温度为35 ℃。显示出与已报道的CMEPA水解酶不同的特性以及较高的催化活性,具有潜在的应用价值。氨基酸序列分析表明DamH具有与酯酶类似的保守序列147GXSXG151和79HGG81,也存在与酯酶活性中心类似的氨基酸残基S149,E244和H274,底物谱实验证实DamH也具有催化对硝基苯酯和三酰甘油酯水解活性[12]。
重组蛋白异源表达的技术瓶颈之一是所表达的蛋白常常以非正确折叠、无活性的包涵体形式存在[13],其解决策略包括包涵体复性,构建新的表达系统,与分子伴侣共表达,引入可溶性分子标签融合表达等[14, 15]。近年来研究者发现氨基酸的突变对蛋白可溶性表达有较大的影响[16, 17],对非活性中心氨基酸残基进行定点突变,逐渐成为提高重组蛋白可溶性表达的策略之一[18]。前期研究中课题组在对DamH进行异源表达时,发现两个非活性位点氨基酸突变(D165G,N192Q)对重组酶可溶性表达和活性都有一定影响(数据未报道)。本研究对其可能的活性中心氨基酸位点(S149,E244和H274)进行定点突变,探索其活性中心氨基酸残基构成。对D165和N192两个氨基酸位点进行定点突变,研究非活性中心氨基酸对重组酶活性及可溶性表达的 影响。
1 材料和方法 1.1 菌株重组酶DamH表达菌株E. coli BL21(DE3)-pET29a-damH,前期构建,保藏于南京农业大学农业部环境微生物与环境工程重点实验室;克隆菌株E. coli DH5α,表达菌株E. coli BL21(DE3),本实验室保藏。
1.2 所用引物本研究所用引物由上海英潍捷基生物技术有限公司(Invitrogen Co.,Ltd.)合成,引物序列见表1。
| DamH gene allele | Mutagenic primer sets |
| Dam-F | CATATGGGCACGTCGCCGCAGTCTGAT |
| Dam-R | GTCGACTCAGTGGTGGTGGTGGTGGTGGGCGCCCTT GAACCACTGGG |
| S149A-F | GCCCGCCGCGTCGCCGCTGGTG |
| S149A-R | CACCAGCGGCGACGCGGCGGGC |
| E244A-F | GCAGGGTCGCGGCACTGCCG |
| E244A-R | CGGCAGTGCCGCGACCCTGC |
| H274A-F | GAAGACGGCCTGCATGCCGT |
| H274A-R | ACGGCATGCAGGCCGTCTTC |
| D165K -F | CTGCGCCAGAAAGGCGTGGCGCT |
| D165K -R | AGCGCCACGCCTTTCTGGCGCAG |
| D165L -F | CTGCGCCAGTTAGGCGTGGCGCT |
| D165L -R | AGCGCCACGCCTAACTGGCGCAG |
| D165E -F | CTGCGCCAGGAAGGCGTGGCGCT |
| D165E -R | AGCGCCACGCCTTCCTGGCGCAG |
| D165P -F | CTGCGCCAGCCTGGCGTGGCGCT |
| D165P -R | AGCGCCACGCCAGGCTGGCGCAG |
| D165Y -F | CTGCGCCAGTATGGCGTGGCGCT |
| D165Y -R | AGCGCCACGCCATACTGGCGCAG |
| D165Q -F | CTGCGCCAGCAAGGCGTGGCGCT |
| D165Q -R | AGCGCCACGCCTTGCTGGCGCAG |
| N192Q -F | ACGCTGCAGACCCAAGCGGCCAC |
| N192Q -R | GTGGCCGCTTGGGTCTGCAGCGT |
| N192K -F | ACGCTGCAGACCAAAGCGGCCAC |
| N192K -R | GTGGCCGCTTTGGTCTGCAGCGT |
| N192L -F | ACGCTGCAGACCTTAGCGGCCAC |
| N192L -R | GTGGCCGCTAAGGTCTGCAGCGT |
| N192E -F | ACGCTGCAGACCGAAGCGGCCAC |
| N192E -R | GTGGCCGCTTCGGTCTGCAGCGT |
| N192P -F | ACGCTGCAGACCCCTGCGGCCAC |
| N192P -R | GTGGCCGCAGGGGTCTGCAGCGT |
| N192Y -F | ACGCTGCAGACCTATGCGGCCAC |
| N192Y -R | GTGGCCGCATAGGTCTGCAGCGT |
| Nucleotide codons in boldface type encode the mutated amino acids. | |
LB培养基(g/L):酵母粉5.0,胰蛋白胨10.0,NaCl 10.0,pH7.0-7.2。
LA-Taq酶,Taq酶,PrimeSTAR HS DNA聚合酶,T4DNA Ligase,pMD19-T Vector均购于大连宝生物公司(TaKaRa),IPTG,X-gal氨苄青霉素(Amp),卡那霉素(Km)购于上海生工,其余各化学试剂均为分析纯。
凝胶回收试剂盒与质粒提取试剂盒购自北京百泰克生物技术有限公司(BioTeke Corporation),核酸测序委托南京金斯瑞生物科技有限公司(Genscript BioTechnologies Co.,Ltd.)。
1.4 重叠延伸PCR定点突变DamH采用重叠延伸的方法,以表达载体质粒pET-29a(+)-damH为模板,设计定点突变引物,对推测的DamH 3个酯酶活性中心位点S149、E244、H274以及D165和N192两个影响重组酶表达和活性的氨基酸位点进行定点突变。
重叠延伸PCR具体操作方法见参考文献[19, 20],以表达载体质粒pET-29a(+)-damH为模板,先分别以全长引物Dam-F(正向引物,含Nde I酶切位点)和突变位点反向引物扩增出上游序列,然后分别以全长引物Dam-R(反向引物,含Sal I酶切位点)和突变位点正向引物扩增出下游序列。PCR产物凝胶电泳,DNA凝胶试剂盒回收后,以两轮扩增的上、下游PCR产物为模板,以全长引物(Dam-F和Dam-R)扩增全长。PCR程序为98 ℃ 10 s,55 ℃ 10 s,72 ℃ 1 min,30个循环;72 ℃ 10 min。
PCR产物凝胶电泳,DNA凝胶试剂盒回收后加A尾,T/A克隆至pMD19-T克隆载体,Nde I/Sal I双酶切后与pET-29a(+)构建突变基因表达载体[21],将表达载体转化至E. coli BL21(DE3)感受态细胞,构建突变表达菌株[22]。
1.5 突变表达菌株诱导表达及突变重组酶的活性测定突变表达菌株静息细胞活性检测:将经0.2 mmol/L IPTG诱导培养后的突变表达菌株培养液6000 r/min,10 min离心,以灭菌的MSM液体培养基重悬洗涤1次,6000 r/min、10 min离心,以灭菌的MSM液体培养基调整OD600至1.0,准确吸取0.1 mL菌悬液至2 mL含2 mmol/L CMEPA的20 mmol/L Tris-HCl 缓冲液(pH6.5)中,35 ℃反应2 h,测活。
1.6 突变重组酶纯化Ni2+-NTA亲和层析柱预先以两倍柱体积的20 mmol/L Tris-HCl(pH7.0)平衡,将含有重组酶DamH的破碎液离心上清上样至Ni2+-NTA亲和层析柱,4 ℃孵育1 h,以5倍柱体积的20 mmol/L Tris-HCl(pH7.0,50 mmol/L咪唑)洗去杂蛋白,然后以20 mmol/L Tris-HCl(pH7.5,300 mmol/L)洗脱,SDS-PAGE电泳[23]检验纯度,收集合并纯度最高的洗脱液,4 ℃,以截留分子量10 kDa的透析袋在20 mmol/L Tris-HCl(pH7.0)缓冲液中透析过夜去除咪唑。
1.7 酶活测定方法取适量纯化的突变重组酶加至3 mL含2 mmol/L CMEPA的20 mmol/L Tris-HCl(pH7.0)中,于30 ℃反应10 min后,加入0.1 mol/L 4-氨基安替比林和0.2 mol/L铁氰化钾各30 μL,以未突变的重组酶(WT)为对照,稀释10倍后测定OD535,根据标准曲线计算MEA含量[12]。
酶活定义为30 ℃下每分钟催化产生1 μmol MEA所需的蛋白量(mg)。
1.8 蛋白含量的测定按Bradford方法进行,以牛血清蛋白作为标准蛋白,具体操作见参考文献[24]。
2 结果和分析 2.1 重叠延伸PCR定点突变DamH以表达载体质粒pET-29a(+)-damH为模板,重叠延伸PCR扩增S149、E244、H274、D165和N192突变基因,结果见图1。
|
| 图1 重叠延伸PCR定点突变damH Figure 1 Agarose gel electrophoresis of site-directed mutagenesis damH by overlapping PCR. M: DL5000 marker; lane 1: damH(WT); lane 2: S149A; lane 3: E149A; lane 4: H149A; lane 5: D165K; lane 6: D165L; lane 7: D165E; lane 8: D165P; lane 9: D165Y; lane 10: N192K; lane 11: N192L; lane 12: N192E; lane 13: N192P; lane 14: N192Y. |
对可能的活性中心氨基酸进行突变后发现,氨基酸的突变影响了酶的稳定性,突变后的表达菌株超声波破碎后检测不到DamH水解活性。以经诱导表达的重组菌株静息细胞进行定性测酶活,结果见图2。显色反应依据MEA在铁氰化钾催化下与4-氨基安替比林形成紫红色物质的原理[12]。

|
| 图2 突变菌株静息细胞测活 Figure 2 CMEPA hydrolysis activity detection of mutant strains. |
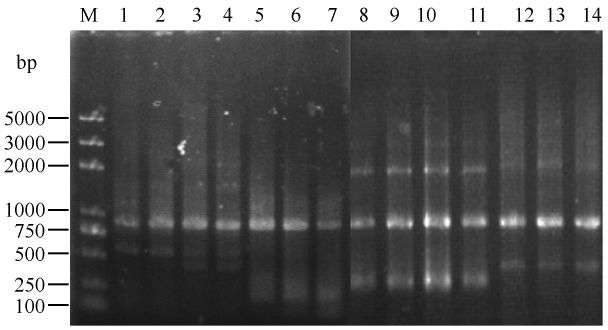
将显色后的反应体系12000 r/min离心1 min,上清液于535 nm波长下测定吸光值,对各菌株的酶活进行比较。与野生型相比,突变株E244A和H274A彻底失活,突变株S149A残留活性为野生型DamH的5%(图2)。洗涤菌体,重悬后超声破碎,12000 r/min离心10 min,取5 μL上清液(约20 μg蛋白),沉淀以少许Tris-甘氨酸缓冲液溶解,取2 μL样品(约4 μg蛋白)进行SDS-PAGE电泳。SDS-PAGE电泳结果显示突变重组蛋白都有一定量的表达,H274A形成较多的包涵体,重组蛋白大量存在于沉淀中(图3)。
|
| 图3 突变表达菌株粗酶液SDS-PAGE电泳图谱 Figure 3 Analysis of the expression of the mutant enzymes on SDS-PAGE. lane 1: total protein of E. coli BL21(DE3) harboring pET-29a(+); lane 2: low molecular protein marker; lane 3: total protein of E. coli BL21(DE3) harboring pET-29a(+)-damH(WT); lane 4: precipitate protein of E. coli BL21(DE3) harboring pET-29a(+)-damH(WT); lane 5: total protein of E. coli BL21(DE3) harboring pET-29a(+)- damH(S149A); lane 6: precipitate protein of E. coli BL21 (DE3) harboring pET-29a(+)-damH(S149A); lane 7: total protein of E. coli BL21(DE3) harboring pET- 29a(+)-damH(E244A); lane 8: precipitate protein of E. coli BL21(DE3) harboring pET-29a(+) -damH(E244A); lane 9: total protein of E. coli BL21(DE3) harboring pET-29a (+)-damH(H274A); lane 10: precipitate protein of E. coli BL21(DE3) harboring pET-29a(+)-damH(H274A). |
将DamH氨基酸序列提交至NCBI 蛋白结构数据库(PDB)进行比对,DamH与酯酶3FAK相似性达30%。以3FAK为参照,将氨基酸序列提交至http://swissmodel.expasy.org/进行结构模拟。将模拟得到的pdb文件以pymol 1.4.2软件绘图,获得DamH蛋白飘带结构和表面形态结构示意图(图4)。与3FAK一致,DamH可能的活性中心3个氨基酸残基S149,E244和H274构成的三联体结构位于由α螺旋和β转角构成的口袋内,N端氨基酸序列构成的α螺旋位于口袋入口处。这一特征与典型的α/β 水解酶家族相符。

|
| 图4 3FAK和DamH晶体结构飘带示意图和表面示意图比较 Figure 4 Compared with monomer and surface structure of 3FAK and DamH. A: monomer structure of 3FAK; B: monomer structure of DamH; C: surface structure of 3FAK; D: surface structure of DamH. |
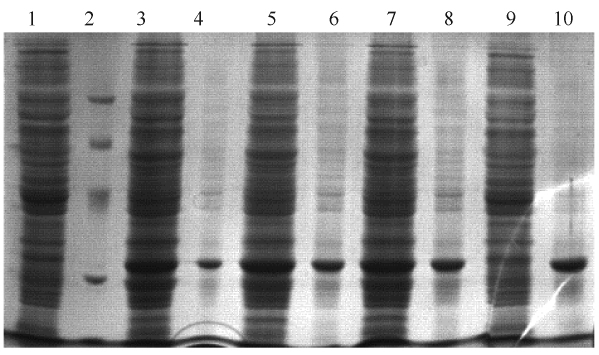
3FAK中与活性相关氨基酸残基(图5-A)中,亲核基团Ser157侧链位置由Trp174稳定,Ser157,Glu251和His281构成电荷中继网,对底物进行共价催化以及酸碱催化。与之类似,DamH可能的活性中心Ser149-His274-Glu244之间也存在类似的结构(图5-B),且与之通过氢键相连的氨基酸残基也与3FAK相似。
|
| 图5 DamH和3FAK活性相关氨基酸残基之间的比较 Figure 5 Reaction related residues of DamH and 3FAK. A: 3FAK; B: DamH structure. H-bond interactions are outlined by the dashed lines. |
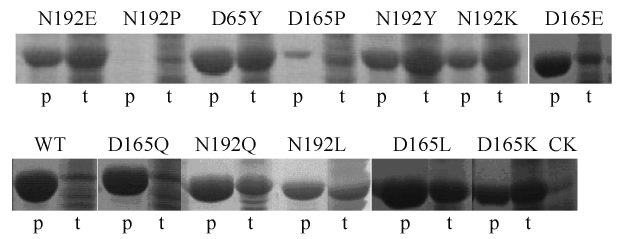
将D165和N192两个氨基酸位点定点突变的重组酶DamH以Ni2+-NTA亲和层析柱进行纯化,破碎上清液取5 μL(约20 μg蛋白),洗脱液取2 μL上样进行SDS-PAGE电泳检测表达情况,结果见图6。
|
| 图6 突变重组酶纯化SDS-PAGE电泳 Figure 6 Analysis of the purification of the mutant enzymes on SDS-PAGE(I). p: purified recombinant DamH; t: total protein of E. coli BL21(DE3) harboring pET-29a(+)-damH; CK: total protein of E. coli BL21(DE3) harboring pET-29a(+) control. |
SDS-PAGE电泳结果显示DamH的2个氨基酸位点突变为脯氨酸后,重组酶表达受到较大的影响,N192P几乎不表达,经Ni2+-NTA亲和层析的洗脱液中未见明显条带;D165P突变株粗酶液中目标蛋白量较少,经Ni2+-NTA亲和层析的洗脱液中有单一条带,但与野生型及其它突变型相比,表达量明显偏低。
2.5 D165和N192位点突变对重组酶酶活的影响将适量经纯化的突变重组酶加入至测活体系,测定CMEPA水解酶活性。同时测定蛋白含量,计算比酶活,以野生型(WT)为对照,结果见表2。由表2可知,非活性氨基酸位点定点突变对DamH可溶性表达有较大影响,突变子N192P和D165P可溶性蛋白表达量分别为野生型表达量的20.8%和28.2%,突变子N192Q、N192K、D165Y、D165E可溶性蛋白表达量分别为野生型表达量的65.6%,64.8%,72.2%和79.5%;突变子N192P、D165P比酶活分别为野生型比酶活的55.5%和49.7%,N192Y和D165Q比酶活分别为野生型的85.5%和82.5%,其余突变株对比酶活无显著影响。
| Mutation | Total protein /mg | Total activity /U | Specific activity /(U/mg) | Purification fold | Yield/% |
| N192Q | 66.28 | 233306 | 3520 | 3.05 | 85.6 |
| N192L | 98.78 | 338519 | 3427 | 3.56 | 86.9 |
| N192K | 65.47 | 218408 | 3336 | 3.01 | 86.3 |
| N192Y | 97.70 | 288117 | 2949 | 3.02 | 84.3 |
| N192P | 21.01 | 40234 | 1915 | 3.12 | 82.5 |
| N192E | 99.93 | 376436 | 3767 | 3.19 | 88.2 |
| D165Q | 98.92 | 281526 | 2846 | 3.14 | 87.4 |
| D165L | 106.96 | 368477 | 3445 | 3.15 | 87.8 |
| D165K | 98.07 | 326965 | 3334 | 3.20 | 84.1 |
| D165Y | 72.95 | 252407 | 3460 | 3.04 | 86.9 |
| D165P | 28.49 | 48860 | 1715 | 3.15 | 83.1 |
| D165E | 80.32 | 282325 | 3515 | 3.06 | 85.3 |
| WT | 101.04 | 348588 | 3450 | 3.13 | 87.7 |
酯酶(EC3.1.1.1)是一类催化酯键水解和形成的酶类,按照氨基酸序列来划分,来源于微生物的酯酶可分为8个家族[25]。家族 VII的酯酶和C族的 β-内酰胺酶具有很高的相似性,它们通常具有Gly-X-Ser-X-Gly保守序列及Ser-X-X-Lys序列。三维结构显示酯酶为α/β水解酶折叠模式,具有一个Ser-Glu(Asp)-His的三联体催化中心,并且活性中心Ser位于Gly-X-Ser-X-Gly序列中[26]。水解蛋白质大分子肽键的丝氨酸蛋白酶家族也具有类似的 Ser-His-Asp催化三联体,在催化机制上也与酯酶类似[27]。
来源于 Rhodococcus sp. N-771的酰胺酶RhAmidase的催化中心由S195-S171-K96构成[28],定点突变结果显示来源于Paracoccus sp. M-1的酰胺酶PamH具有与之类似的活性中心氨基酸残基(K84,S159,S183)[10]。通过氨基酸序列比较,DamH并不存在与PamH类似的三联体结构。对DamH可能的3个活性中心氨基酸进行定点突变后,获得的S149A的芳基酰胺酶活性只有野生型的5%,而突变型E244A和H274A的活性则完全丧失,表明这3个氨基酸与催化作用密切相关。Nam、Palm、Zhu等通过X-ray衍射分别解析了酯酶EstE5、PestE、EstE7、HerE的晶体结构,底物共结晶试验结果表明这些酯酶的活性中心都由Ser-Glu(Asp)-His三联体结构组成[29, 30, 31, 32]。本研究中定点突变的结果揭示DamH的活性中心由Ser-Glu-His构成,与酯酶类似。其组成形式与已报道的CMEPA水解酶PamH、AmpA及RhAmidase完全不同。
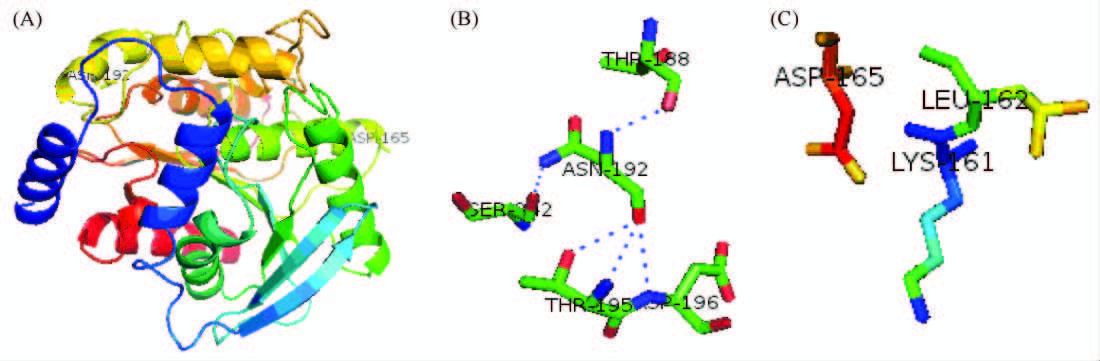
蛋白质理性设计通常用于优化酶的稳定性、活性及底物对映体选择性等[33],大量的实验证据显示定向进化可有效提高靶蛋白的可溶性表达[17, 18, 34]。影响重组蛋白可溶性表达的因素包括亲水性残基的构成、等电点等。突变对DamH可溶性表达的影响可以通过同源结构建模获得部分解释。DamH的D165残基和N192残基均位于DamH球状蛋白的表面(图7-A),且位于两个α螺旋的转角处,这一位置氨基酸残基的改变可能对构象的维持有重要作用。与D165相互作用的氨基酸为Lys161和Leu162(图7-C),与N192作用的氨基酸为Thr188,Thr195,Asp196及Ser242(图7-B)。本研究结果显示两个极性氨基酸D165、N192突变为非极性氨基酸Pro,以及第192位不带电的Asp突变为碱性氨基酸Lys后,重组酶可溶性表达受到影响最大。可能是因为脯氨酸的芳香环侧链这种刚性结构对蛋白的正确折叠及维持构象的稳定性产生一定的影响,从而导致蛋白的可溶性表达和比酶活的差异。
|
| 图7 N192和D165在DamH上所处位置及相关氨基酸残基 Figure 7 The N192 reaction related residues and location on the DamH. |
| [1] | Hou Y, Dong W, Wang F, Li J, Shen W, Li Y, Cui Z. Degradation of acetochlor by a bacterial consortium of Rhodococcus sp. T3-1, Delftia sp. T3-6 and Sphingobium sp. MEA3-1. Letters in Applied Microbiology, 2014, 59(1): 35-42. |
| [2] | Hou Y, Wang F, Dong WL, Cui ZL. Degradation characteristics of all acetochlor-degrading bacterium Rhodococcus sp. T3-l. China Environmental Science, 2013, 33(10): 1785-1790. (in Chinese) 侯颖, 王飞, 董维亮, 崔中利. Rhodococcus sp. T3-1 菌株降解乙草胺的特性. 中国环境科学, 2013, 33(10): 1785-1790. |
| [3] | Xiao NW, Jing BB, Ge F, Liu XH. The fate of herbicide acetochlor and its toxicity to Eisenia fetida under laboratory conditions. Chemosphere, 2006, 62(8): 1366-1373. |
| [4] | Coleman S, Linderman R, Hodgson E, Rose RL. Comparative metabolism of chloroacetamide herbicides and selected metabolites in human and rat liver microsomes. Environmental Health Perspectives, 2000, 108(12): 1151-1157. |
| [5] | Muthukaruppan G, Janardhanan S, Vijayalakshmi G. Sublethal toxicity of the herbicide butachlor on the earthworm Perionyx sansibaricus and its histological changes (5 pp). Journal of Soils and Sediments, 2005, 5(2): 82-86. |
| [6] | Geng BR, Yao D, Xue QQ. Acute toxicity of the pesticide dichlorvos and the herbicide butachlor to tadpoles of four anuran species. Bulletin of Environmental Contamination and Toxicology, 2005, 75(2): 343-349. |
| [7] | Panneerselvam N, Sinha S, Shanmugam G. Butachlor is cytotoxic and clastogenic and induces apoptosis in mammalian cells. Indian Journal of Experimental Biology, 1999, 37(9): 888-892. |
| [8] | Li Y, Chen Q, Wang CH, Cai S, He J, Huang X, Li SP. Degradation of acetochlor by consortium of two bacterial strains and cloning of a novel amidase gene involved in acetochlor-degrading pathway. Bioresource Technology, 2013, 148: 628-631. |
| [9] | 侯颖. 芳氧苯氧丙酸类和氯乙酰胺类除草剂降解菌的分离鉴定、降解基因克隆表达及其代谢途径研究. 南京农业大学博士学位论文, 2011. |
| [10] | Shen WL, Chen HH, Jia KZ, Ni J, Yan X, Li SP. Cloning and characterization of a novel amidase from Paracoccus sp. M-1, showing aryl acylamidase and acyl transferase activities. Applied Microbiology and Biotechnology, 2012, 94(4): 1007-1018. |
| [11] | Zhang J, Yin JG, Hang BJ, Cai S, He J, Zhou SG, Li SP. Cloning of a novel arylamidase gene from Paracoccus sp. strain FLN-7 that hydrolyzes amide pesticides. Applied and Environmental Microbiology, 2012, 78(14): 4848-4855. |
| [12] | Wang F, Hou Y, Zhou J, Li ZK, Huang Y, Cui ZL. Purification of an amide hydrolase DamH from Delftia sp. T3-6 and its gene cloning, expression, and biochemical characterization. Applied Microbiology and Biotechnology, 2014, 98(17): 7491-7499. |
| [13] | Wetzel R. For protein misassembly, it’s the “I” decade. Cell, 1996, 86(5): 699-702. |
| [14] | Zhang YB, Howitt J, McCorkle S, Lawrence P, Springer K, Freimuth P. Protein aggregation during overexpression limited by peptide extensions with large net negative charge. Protein Expression and Purification, 2004, 36(2): 207-216. |
| [15] | Liu S, Hu BC. Strategy of protein soluble expression in Escherichia coli. Letters in Biotechnology, 2005, 16(2): 172-175. (in Chinese) 刘爽, 胡宝成. 原核系统可溶性表达策略. 生物技术通讯, 2005, 16(2): 172-175. |
| [16] | Dale GE, Broger C, Langen H, D'Arcy A, Stüber D. Improving protein solubility through rationally designed amino acid replacements: solubilization of the trimethoprim-resistant type S1 dihydrofolate reductase. Protein Engineering, 1994, 7(7): 933-939. |
| [17] | Yang JK, Park MS, Waldo GS, Suh SW. Directed evolution approach to a structural genomics project: Rv2002 from Mycobacterium tuberculosis. Proceedings of the National Academy of Sciences of the United States of America, 2003, 100(2): 455-460. |
| [18] | Jiang SM, Li CH, Zhang WW, Cai YH, Yang YL, Yang S, Jiang WH. Directed evolution and structural analysis of N-carbamoyl-D-amino acid amidohydrolase provide insights into recombinant protein solubility in Escherichia coli. Biochemical Journal, 2007, 402(3): 429-437. |
| [19] | Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene, 1989, 77(1): 51-59. |
| [20] | Landt O, Grunert HP, Hahn U. A general method for rapid site-directed mutagenesis using the polymerase chain reaction. Gene, 1990, 96(1): 125-128. |
| [21] | Sambrook J, Russell D. Molecular cloning, a laboratory manual. Vol. 3. New York: Cold Spring Harbor Laboratory Press, 2001. |
| [22] | Hou Y, Tao J, Shen WJ, Liu J, Li JQ, Li YF, Cao H, Cui ZL. Isolation of the fenoxaprop-ethyl (FE)-degrading bacterium Rhodococcus sp. T1, and cloning of FE hydrolase gene feh. FEMS Microbiology Letters, 2011, 323(2): 196-203. |
| [23] | Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature, 1970, 227(5259): 680-685. |
| [24] | Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Analytical Biochemistry, 1976, 72(1): 248-254. |
| [25] | Gupta R, Gupta N, Rathi P. Bacterial lipases: an overview of production, purification and biochemical properties. Applied Microbiology and Biotechnology, 2004, 64(6): 763-781. |
| [26] | Petersen EI, Valinger G, Slkner B, Stubenrauch G, Schwab H. A novel esterase from Burkholderia gladioli which shows high deacetylation activity on cephalosporins is related to β-lactamases and dd-peptidases. Journal of Biotechnology, 2001, 89(1): 11-25. |
| [27] | Sun ZR, Wang Y, Hu SM, Guo Q, He FC. Study on the molecule structural evolution of serine proteinase superfamily. Acta Biophysica Sinica, 1999, 15(3): 530-535. (in Chinese) 孙之荣, 王钰, 胡胜民, 郭青, 贺福初. 丝氨酸蛋白酶超家族分子结构进化研究. 生物物理学报, 1999, 15(3): 530-535. |
| [28] | Ohtaki A, Murata K, Sato Y, Noguchi K, Miyatake H, Dohmae N, Yamada K, Yohda M, Odaka M. Structure and characterization of amidase from Rhodococcus sp. N-771: Insight into the molecular mechanism of substrate recognition. Biochimica et Biophysica Acta (BBA)-Proteins and Proteomics, 2010, 1804(1): 184-192. |
| [29] | Nam KH, Kim MY, Kim SJ, Priyadarshi A, Lee WH, Hwang KY. Structural and functional analysis of a novel EstE5 belonging to the subfamily of hormone-sensitive lipase. Biochemical and Biophysical Research Communications, 2009, 379(2): 553-556. |
| [30] | Nam KH, Kim MY, Kim SJ, Priyadarshi A, Kwon ST, Koo BS, Yoon SH, Hwang KY. Structural and functional analysis of a novel hormone-sensitive lipase from a metagenome library. Proteins: Structure, Function, and Bioinformatics, 2009, 74(4): 1036-1040. |
| [31] | Palm GJ, Fernández-lvaro E, Bogdanovi Dc' X, Bartsch S, Sczodrok J, Singh RK, Bttcher D, Atomi H, Bornscheuer UT, Hinrichs W. The crystal structure of an esterase from the hyperthermophilic microorganism Pyrobaculum calidifontis VA1 explains its enantioselectivity. Applied Microbiology and Biotechnology, 2011, 91(4): 1061-1072. |
| [32] | Zhu XY, Larsen NA, Basran A, Bruce NC, Wilson IA. Observation of an arsenic adduct in an acetyl esterase crystal structure. Journal of Biological Chemistry, 2003, 278(3): 2008-2014. |
| [33] | Zhu L, Zhou Q, Zhan BJ, Chi LY Zhang JL. Progress and applications of site-directed mutagenesis of lipases and esterases. Strait Pharmaceutical Journal, 2012, 24(2): 10-13. (in Chinese) 祝玲, 周琼, 詹冰津, 池丽影, 张军玲. 脂肪酶及酯酶定点突变研究进展及其应用. 海峡药学, 2012, 24(2): 10-13. |
| [34] | Wigley WC, Stidham RD, Smith NM, Hunt JF, Thomas PJ. Protein solubility and folding monitored in vivo by structural complementation of a genetic marker protein. Nature Biotechnology, 2001, 19(2): 131-136. |
 2015, Vol. 55
2015, Vol. 55