中国科学院微生物研究所,中国微生物学会,中国菌物学会
文章信息
- 麻婷婷, 承磊, 刘来雁, 代莉蓉, 周正, 张辉. 2015
- Tingting Ma, Lei Cheng, Laiyan Liu, Lirong Dai, Zheng Zhou, Hui Zhang. 2015
- 不同抑制剂对乙酸降解产甲烷及产甲烷菌群结构的影响
- Effects of selective methanogenic inhibitors on methanogenesis and methanogenic communities in acetate degrading cultures
- 微生物学报, 2015, 55(5): 587-597
- Acta Microbiologica Sinica, 2015, 55(5): 587-597
-
文章历史
- 收稿日期:2014-10-20
- 修回日期:2014-12-25
乙酸是沼气发酵过程中重要的中间代谢产物,其进一步转化为甲烷主要通过2种途径,一是乙酸营养型产甲烷菌的直接裂解产生甲烷[1],这主要由乙酸营养型产甲烷古菌甲烷鬃毛菌属(Methanosaeta)和甲烷八叠球菌属(Methanosarcina)完成[2]。另一种是通过乙酸互营氧化途径来完成,热力学分析表明乙酸转化为H2和CO2在标准状况下是无法自发进行的,因为这是吸热反应[△G0′>0,表1中反应(2)][3]。但是当这个反应和产甲烷代谢偶联后,乙酸降解产生的H2迅速被氢营养型产甲烷古菌利用,整个反应体系中保持着极低的氢分压,那么乙酸降解产生H2和CO2的反应就可以连续进行[表1中反应(3)],这也表明乙酸互营氧化产甲烷代谢需要两类不同的微生物通过互营代谢作用来完成。第一步是乙酸被互营菌氧化为H2和CO2,紧接着氢营养型产甲烷菌利用H2还原CO2为CH4。目前已报道的互营乙酸氧化菌有Clostridium ultunense[4]、Syntrophaceticus schinkii[5]、Tepidanaerobacter acetatoxydans[6]、Thermacetogenium phaeum[7]、Thermotoga lettingae[8]和已经丢失的AOR菌株[9]。基于微生物分子生态学和同位素示踪等手段发现在沼气发酵过程中普遍存在乙酸互营氧化产甲烷代谢过程[10, 11]。
| Process | Reaction | △G0′(KJ/mol) |
| *labeled carbon | ||
| (1)Aceticlastic methanogenesis | *CH3COO-+ H2O → *CH4+ HCO3- | -31.0 |
| (2)Syntrophic acetate oxidation | *CH3COO-+ 4H2O → H*CO3-+4H2+ HCO3-+H+ | +104.6 |
| (3)Hydrogenotrophic methanogenesis | 4H2+ HCO3-+ H+→ CH4+ 3H2O | -135.6 |
| (4)2+3 | *CH3COO-+H2O → H*CO3-+ CH4 | -31.0 |
| (5)Homoacetogenesis | 4H2+HCO3-+ H+→ CH3COO-+4H2O | -104.6 |
石油烃厌氧生物降解产甲烷的微生物学过程和机理研究是近年来石油微生物领域研究的热点之一,在残余油藏开发和原位厌氧生物修复等领域具有广泛的应用前景[13, 14]。乙酸也是石油烃降解产甲烷过程中的重要中间代谢产物,以正十六烷烃为例,热力学分析表明其降解产甲烷过程存在五种可能的产甲烷代谢过程[15]:(i)互营降解烃产生H2和CO2,再通过CO2还原作用(氢营养型产甲烷古菌)产生CH4,(ii)互营降解烃产生H2和乙酸,通过CO2还原和乙酸裂解作用产生CH4(分别是氢营养型和乙酸营养型产甲烷古菌),(iii)互营降解烃产生H2和乙酸,通过乙酸互营氧化作用,转化乙酸产生H2和CO2,最后通过CO2还原作用产生CH4,(iv)互营降解烃产生乙酸,通过乙酸裂解作用产生CH4,(v)互营降解烃产生乙酸,通过乙酸互营氧化转化为H2和CO2,最后通过CO2还原作用产生CH4。热力学分析发现温度会影响石油烃降解产甲烷途径,而且不同营养类型产甲烷古菌在不同温度条件下的生长动力学存在较大差异[15, 16]。
目前国内外多个小组富集获得了不同温度条件下的烷烃降解产甲烷菌系[17, 18, 19, 20, 21, 22],发现了不同的微生物群落结构组成。如Widdel小组发现在正十六烷烃降解产甲烷富集物(28-30℃)中古菌主要是由乙酸营养型产甲烷古菌Methanosaeta、氢营养型产甲烷古菌Methanoculleus和Methanospirillum组成[13]。Larter小组发现室温条件下富集获得的原油降解产甲烷菌系中主要是氢营养型产甲烷古菌为主[23],Suflita小组发现原油降解产甲烷富集物(35℃)中所有古菌克隆都属于Methanosaeta[18],后来他们又发现在高温原油降解产甲烷条件下(55℃)的古菌群落主要由氢营养型产甲烷古菌Methanothermobacter组成[24]。Mbadinga等[25]也发现Methanothermobacter是高温饱和烃降解产甲烷富集物中的优势古菌类群之一。我们小组从胜利油田同一采集点富集获得3个不同温度的石油烃降解产甲烷富集物Y15(15℃)、M82(35℃)和SK(55℃),发现不同温度条件下的产甲烷古菌类群也是不同的[26, 27](15℃的文章正在审稿中)。这些研究表明在石油烃降解产甲烷过程中可能存在多种不同的产甲烷代谢途径,但是迄今为止,还没有看到定性定量研究石油烃降解产甲烷代谢途径的研究报道。
由于沼气发酵过程的复杂性,研究产甲烷代谢途径的方法主要有稳定同位素示踪结合微生物分子生态学技术[28]、同位素自然分馏[29]和添加不同类型产甲烷古菌的选择性抑制剂等。其中NH4Cl[30]和CH3F[31]可以选择性抑制乙酸营养型产甲烷古菌,以阻断或者削弱乙酸裂解产甲烷途径。本研究采用从胜利油田富集获得的3个不同温度条件下的正十六烷烃降解产甲烷富集物Y15(15℃)、M82(35℃)和SK(55℃)作为接种物,分别添加NH4Cl和CH3F,并以乙酸为唯一碳源进行传代培养。定期监测甲烷产生趋势,并结合末端限制性片段多态性(T-RFLP)和16S rRNA基因克隆文库,分析乙酸降解产甲烷趋势和产甲烷古菌的菌群结构,研究不同温度条件下乙酸降解产甲烷的可能代谢途径,这将为研究石油烃降解产甲烷途径和代谢机理提供指导,并为开展石油微生物技术的应用提供理论支撑。
1 材料和方法 1.1 富集培养本实验接种物来自课题组富集驯化的正十六烷烃降解产甲烷菌系Y15(15℃)、M82(35℃)和SK(55℃)[22, 26, 27]。利用Hungate厌氧操作技术配制厌氧无机盐培养基[32, 33]。共设置5组,其中Ac(+)组只加NaAc(820.3 mg/L),AC+NH4Cl组加NH4Cl(3g/L)和NaAC(820.3mg/L),AC+CH3F组中加入CH3F(1.5%)和NaAC(820.3mg/L),Ac(-)组不加NaAc和抑制剂。接种量为20%(V/V),每组3个重复,用HCl(1 mol/L)或NaOH(1 mol/L)调pH为7.0-7.5,分别于15℃、35℃和55℃条件下静置暗培养。
1.2 化学分析方法采用气相色谱GC-2010(Shimadzu、日本)测定气体组分与含量[33]。
采用气相色谱7890A(Agilent,美国)测定乙酸浓度[33]。
1.3 微生物分子分子生态学分析1.3.1 基因组DNA的提取及纯化具体操作参见文献[34]。
1.3.2 PCR扩增及纯化具体操作参见文献[33]。
1.3.3 酶切及产物纯化:用紫外/可见分光光度计(Beckman、德国)测量PCR产物浓度。其中15℃和35℃古菌PCR产物使用Taq I(TaKaRa,日本)65℃酶切3.5 h;55℃古菌PCR产物使用AluI(TaKaRa,日本)37℃酶切3.5 h。其余操作参见文献[33, 34]。
1.3.4 T-RFLP分析其余操作参见文献[33, 34]。
1.3.5 16S rRNA基因克隆文库的构建、测序:除了引物没有使用荧光标记以外,PCR体系和扩增程序和上述T-RFLP分析一样。切胶纯化回收PCR产物,并连接到质粒pMDTM19-T(TaKaRa、日本),然后导入大肠杆菌感受态细胞DH-5α(Tiangen、中国)。挑出白色单克隆并涂布LB固体培养基上,37℃过夜培养后提取质粒DNA后测序(ABI、美国)。使用greengene数据库中的“Chimera check with Bellerophon”进行嵌合体检验[35],使用Mothur将相似性97%以上的序列作为一个分类操作单元(OTU)进行聚类[36],所有序列上传到RDP数据库,采用Classifier程序确定克隆文库中序列的分类地位,从每个OTU中选取一个代表序列,在RDP数据库中使用seqmatch程序查找其最相似菌株[37]。根据Good’s formula计算构建克隆文库多样性的覆盖度[38]。所有16S rRNA基因序列在GenBank中的登录代码为KJ735836-KJ735838、KJ735842-KJ735843、KJ735847-KJ735848、KJ744107-KJ744147和KJ744170-KJ744200。
1.4 统计分析不同抑制剂对乙酸降解产甲烷菌影响的甲烷最大产量和最大甲烷比生长速率采用单因素方差分析(SPSS 16.0、美国)。
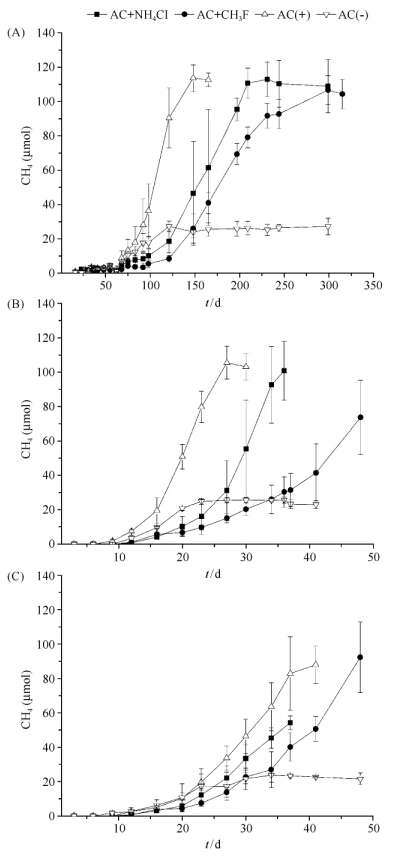
2 结果和分析 2.1 不同温度条件下中抑制剂对乙酸降解产甲烷趋势的影响添加乙酸传代培养,菌系Y15、M82和SK都可以在对应的温度条件下产生甲烷。但是乙酸降解产甲烷速率不一样。在15℃条件下菌系Y15利用乙酸的生长周期约为150d(图1-A),在35℃条件下菌系M82降解乙酸的周期约为30d(图1-B),在55℃条件下菌系SK降解乙酸的周期约为40d(图1-C)。添加抑制剂NH4Cl和CH3F后,不同温度条件下乙酸产甲烷的延滞期增加,甲烷产生速率下降(图1、表2)。其中15℃条件下,不添加抑制剂的AC(+)组的最大比生长速率是0.067±0.013 d-1,显著高于AC+CH3F组的0.036±0.003 d-1(p≤ 0.05),也高于AC+NH4Cl组的0.056±0.014 d-1(p> 0.05)。另外AC+NH4Cl组的最大比生长速率要高于AC+CH3F组(p> 0.05)。35℃条件下,不添加抑制剂的AC(+)组最大比生长速率是0.376±0.011 d-1 AC+NH4Cl组0.269±0.014 d-1(p≤ 0.01)和显著高于AC+CH3F组0.113±0.035 d-1(p<0.01)。另外AC+NH4Cl组的最大比生长速率要高于AC+CH3F组(p<0.01)。55℃条件下,不添加抑制剂的AC(+)组最大比生长速率是0.244±0.014 d-1,显著高于AC+NH4Cl组的0.226±0.041 d-1(p<0.05)和AC+CH3F组的0.121±0.012 d-1(p≤ 0.01)。此外AC+NH4Cl组的最大比生长速率要高于AC+CH3F组(p<0.05)。
| T/℃ | Experimental group | Incubation time/d | Maximum specific growth rate (d-1) | c(Acetate)/(mg/L) |
| ND: Acetate concentration below the instrument detection limit(<10 mg/L). AC(-): the cultures without aceate addition, AC(+): the cultures amended with aceate,AC+NH4Cl: the cultures amended with acetate and NH4Cl, AC+CH3F: the cultures amended with acetate and CH3F. | ||||
| 15 | AC(-) | 300 | - | ND |
| AC(+) | 165 | 0.067±0.013 | 20.3±4.3 | |
| AC+NH4Cl | 300 | 0.056±0.014 | ND | |
| AC+CH3F | 315 | 0.036±0.003 | ND | |
| 35 | AC(-) | 48 | - | ND |
| AC(+) | 30 | 0.376±0.011 | ND | |
| AC+NH4Cl | 36 | 0.269±0.014 | 67.7±15.3 | |
| AC+CH3F | 48 | 0.113±0.035 | 165.6±30.5 | |
| 55 | AC(-) | 48 | - | ND |
| AC(+) | 41 | 0.244±0.014 | 39.5±15.6 | |
| AC+NH4Cl | 38 | 0.226±0.041 | 287.9±13.8 | |
| AC+CH3F | 48 | 0.121±0.012 | 115.4±42.4 | |
|
| 图1. 不同温度条件下乙酸降解产甲烷趋势图 Figure 1. Time course of methane production form cultures incubated at different tempertures. A: 15℃; B: 35℃; C: 55℃. |
2.2.1 低温条件下乙酸降解产甲烷菌系的古菌群落结构:T-RFLP分析发现15℃条件下,添加不同抑制剂之间的古菌群落组成相似,主要是由片段(T-RFs)186 bp和284 bp组成(图2-A)。其中T-RF 284 bp是丰度最高的片段,在AC(-)、AC(+)和AC+NH4Cl组中丰度高达93%-99%,在AC+CH3F组中丰度偏低,但仍然有77.1±14.5%(图2-A)。T-RF 186 bp在AC+CH3F组中丰度最高为15.7±20.1%,在AC(+)组中丰度为4.3±4.3%,在AC(-)组和AC+NH4Cl组中丰度为1.2±2.1%和0.2±0.4%。结合古菌16S rRNA基因克隆文库分析(表3),发现T-RF 284 bp代表的克隆与Methanosaeta conciliiGP6T的相似性是100%(表3)。M. conciliiGP6T只能利用乙酸作为唯一碳源并产生甲烷,其温度生长范围在10-45℃之间,最适生长温度35-40℃[39, 40]。T-RF 186 bp代表的古菌克隆与Methanosarcina mazei S-6T的相似性最高也是100%[41, 42, 43, 44],与最近从西伯利亚冻土中分离的Methanosarcina soligelidi SMA-21T的相似性也高达99%[45]。M. mazei不同菌株的生理生化特征差异较大,如模式菌株S-6T可以利用乙酸和甲基类化合物,但不利用H2/CO2生长产CH4,最适生长范围在30-40℃,最低生长温度可能小于20℃[41, 42, 43, 44],而菌株LYC除了可以甲基类化合物生长,还可以利用H2/CO2,但是不利用乙酸生长产甲烷[46]。M. soligelidiSMA-21T可以利用乙酸、甲醇和H2/CO2等3种不同类型的碳源产甲烷,并且温度生长范围宽泛(0-54℃)[45]。

|
| 图2. 低温(A)、中温(B)和高温(C)条件下乙酸降解产甲烷菌系的古菌T-RFLP图 Figure 2. The archaeal T-RFLP profile of methanogenic acetate-degrading cultures incubated at 15℃(A), 35℃(B) and 55℃(C). |
2.2.2 中温条件下乙酸降解产甲烷菌系的古菌群落结构:T-RFLP分析发现35℃条件下,添加不同抑制剂之间的古菌群落主要是由T-RFs 186 bp、228 bp和495 bp组成(图2-B)。T-RF 495 bp在所有处理组中的丰度都是最高的,其中在AC(+)组中丰度高达95.1±2.3%,在AC(-)和AC+NH4Cl组中丰度为77-79%,在AC+CH3F组中丰度最低,也达到了44.4±11.7%。T-RF 228 bp在AC+CH3F组中为优势菌群丰度为42.0±6.1%,在其他3组中的丰度范围是2%-8%。T-RF 186 bp在AC(+)组中丰度最低为2.6±1%,在其他3组中的丰度为11%-14%。结合克隆文库(表3)分析发现,T-RF 495 bp代表的克隆与Methanosaeta harundinacea8AcT相似性最高为96%,M. harundinacea8AcT是一种杆状的乙酸营养型产甲烷古菌,利用乙酸作为碳源进行生长产CH4,不能利用H2/CO2、甲酸、甲醇和乙醇等作为碳源进行生长,其最适生长温度在34-37℃[47]。T-RF 186 bp代表的克隆与Methanoculleus receptaculiZC-2T的相似性为99%,M. receptaculiZC-2T分离自胜利油田,最适生长温度在50-55℃,但是可在30℃以下生长,只能利用H2/CO2产生CH4[48]。本研究中T-RF 228 bp所代表的古菌类群没有检测到,但是根据我们之前的研究发现T-RF 228 bp代表的是乙酸营养型产甲烷古菌Methanosaetasp.[22]。
2.2.3 高温条件下乙酸降解产甲烷菌系的古菌群落结构:T-RFLP分析发现55℃条件下,添加不同抑制剂之间的古菌群落组成相似,主要是由T-RFs 68 bp和169 bp组成(图2-C),但是相对丰度差别较大。其中T-RF 68 bp在AC+NH4Cl和CH3F组中丰度为48.5-84.3%,明显高于AC(+)组的8.8±3.4%。T-RF 169 bp在AC(+)组中丰度最高为82.8±9.0%,而在AC+CH3F组和AC+NH4Cl组中丰度分别只有35.8±7.8%和6.1±8.6%。结合16S rRNA基因克隆文库分析(表3),发现T-RF 68 bp代表的克隆与Methanothermobacter crinaleTm2T相似度最高为99%,菌株Tm2T分离自胜利油田,只能利用H2/CO2生长并产生甲烷,最适生长温度为65℃[49]。T-RF 169 bp代表的克隆与Methanosaeta thermophila的相似度最高为98%。M. thermophila分离自厌氧反应器中,只能裂解乙酸产甲烷,而不能利用H2/CO2和甲基类碳源生长,最适生长温度为55-60℃[50]。
| Phylogenetic group | In silicoT-RF (No. of clone)# | Type clone (GenBank) | The most similar species (similarity) | ||||
| 15-AC | 15-F | 35-NH | 35-AC | 55-F | |||
| Remark:The clones (total 79 sequences) were assigned and grouped on the sequence similarity of 97%, each column represents different type clone OTU.Number in the parentheses indicates numbers of clones.15-AC: clones retrieved from acetate culture incubated at 15℃;15-F: clones retrieved from acetate culture incubated at 15℃ with CH3F;35-NH: clones retrieved from acetate culture incubated at 35℃ with NH4Cl;35-AC: clones retrieved from acetate culture incubated at 35℃;55-F: clones retrieved from acetate culture incubated at 15℃ with CH3F. | |||||||
| Methanosaetaceae | 283(1) 284(10) 285(1) | 284(2) | A156-4(KJ735837) | Methanosaeta concilii; NR102903 (100%) | |||
| 495 (11) | 495 (12) | A56-8 (KJ735843) | Methanosaeta harundinacea; NR102896 (96%) | ||||
| 169 (5) | A48-7(KJ735842) | Methanosaeta thermophila; NR_074214 (98%) | |||||
| Methanosarcinaceae | 186(4) | 186(14) | A156-11 (KJ735836) | Methanosarcinamazei; CP004144 (100%) | |||
| Methanobacteriaceae | 67(1) 68(11) | A48-18 (KJ735838) | Methanothermobacter crinale; HQ283273 (99%) | ||||
| 473(1) | A48-17 (KJ735848) | Methanocaldococcus vulcanius; NR_074195 (76%) | |||||
| Thermofilaceae | 31(1) | A48-11 (KJ735847) | Thermofilum pendens; NR074406 (83%) | ||||
| Methanomicrobiaceae | 186(1) | 186(2) | A44-7 (KJ744109) | Methanoculleus receptaculi; DQ787475 (99%) | |||
石油烃降解产甲烷过程存在5种可能的产甲烷代谢途径,其中温度是影响产甲烷代谢途径的重要因素之一[15]。石油烃降解产生H2/CO2、H2和乙酸的吉布斯自由能(△G)随温度升高而降低,这在热力学上是有利的。与此不同,石油烃降解产生乙酸的△G随温度升高呈增大趋势[15]。另外,在产甲烷过程中,虽然CO2还原产CH4和乙酸裂解产甲烷过程都是放热过程(△G<0),可以自发进行,但是利用H2还原CO2产CH4的反应的△G随温度上升下降,而乙酸裂解产CH4呈继续下降趋势[51]。此外,乙酸氧化产H2和CO2在标准状况下的△G>0,但是这个反应的自由能随温度上升而下降[51]。本小组之前从胜利油田某油泥沙富集获得了3个不同温度条件(15℃、35℃、55℃)的石油烃降解产甲烷菌系Y15、M82和SK。为了研究石油烃降解产甲烷的代谢途径及其产甲烷古菌菌群结构,我们直接添加乙酸并进行传代培养,结合T-RFLP和克隆技术这些经典的微生物群落组成与演替规律的分析方法[52]。本研究发现这3个菌系都可以利用乙酸生长并产生甲烷,进一步分析发现降解乙酸的古菌群落中,乙酸营养型产甲烷古菌Methanosaeta占绝对优势,这表明在石油烃降解产甲烷菌系中,可能都存在裂解乙酸产甲烷代谢途径,并且参与乙酸裂解的产甲烷古菌属于Methanosaeta属下的3个不同种,其中15℃条件下降解乙酸的是耐冷的M. concilii,35℃条件下主要是嗜中温的M.harundinacea,而在55℃条件下主要是嗜热的M.thermophila,这也表明油泥沙中存在丰富多样的乙酸营养型产甲烷古菌。
采用选择性抑制剂CH3F和NH4Cl来特异性抑制乙酸营养型产甲烷古菌是验证产甲烷代谢途径的重要方法之一[53, 54]。Frenzel等报道CH3F不仅可以抑制甲烷氧化过程,还可以选择性抑制乙酸裂解产甲烷的活性[55, 56]。Conrad等发现添加0.01%-1%的CH3F就可以抑制乙酸裂解产甲烷活性,当CH3F浓度过高时,对CO2还原产CH4代谢也会有一定的影响[53]。他们后来还发现CH3F不仅会抑制Methanosaeta的产甲烷活性,还会抑制Methanosarcina的生长[57]。我们的研究发现添加CH3F会增大3个菌系的产甲烷延滞期,但是在低温条件下,CH3F增高了M. mazei相关的古菌丰度,M. mazei也可以利用乙酸生长产甲烷[41],这表明低温条件下添加CH3F抑制剂后,代谢过程可能主要还是通过直接裂解乙酸产生甲烷的。在中温条件下M. harundinacea相关的古菌丰度降低,但是另外一类乙酸营养型产甲烷古菌的Methanosaeta sp.和氢营养型产甲烷古菌M. receptaculi丰度增加,这表明有一部分乙酸可能被氧化为H2和CO2,然后通过M. receptaculi转化为CH4,另外有部分乙酸通过两种不同的Methanosaeta裂解产生乙酸。这些发现也表明CH3F对不同种属乙酸营养型产甲烷古菌活性的抑制是不同的。在高温条件下,氢营养型产甲烷古菌M. crinale丰度相对于中低温增幅最明显,前期研究发现M. crinale相关的古菌类群广泛分布在油藏环境中,在高温原油降解产甲烷和油藏乙酸互营氧化中起着重要的生理生态学功能[24]。与CH3F类似,NH4Cl也会影响产甲烷古菌,特别是乙酸营养型产甲烷古菌的产甲烷活性[58, 59, 60, 61]。文献报道显示氨浓度在1.7-14g/L会对产甲烷过程产生抑制,氨主要从浓度、pH和菌体的适应性几个方面对产甲烷过程造成影响[62]。目前推测存在多种可能的NH4+抑制机理,一种认为NH4+溶解在水中形成的氨(NH3)会造成产甲烷古菌细胞内的钾流失,并破坏胞内pH平衡[61, 63],另外有学者提出NH3会抑制胞内与产甲烷代谢相关的酶[64],Zhang等认为NH4+抑制了产甲烷代谢的转录表达活性[65]。Fotidis等认为NH4+增加了维持反应进行下去的能量,消耗细胞内的K+而且还抑制了特定的酶反应[66]。但是NH4+对Methanosaeta的抑制作用要明显高于其他类型产甲烷古菌的原因还不清楚。另外,Steinhaus等发现M. concilii利用NH4-N最适生长浓度为0.25-1.1 g/L(pH 7.6)[67],但Sprott等发现0.56 g/L NH4-N会完全抑制M. concilii的产甲烷活性(pH 7.0)[59]。在不同的沼气发酵反应器中,抑制Methanosaetaceae产甲烷活性的NH4-N浓度波动范围也比较大(1.5-7 g/L)[30, 68, 69, 70]。
在本研究中,添加同样浓度的NH4-N培养,发现从低温到高温条件下,Methanosaeta相对丰度明显降低,而氢营养型产甲烷古菌丰度显著上升,这与之前的研究发现类似[70],这可能和接种物、培养条件和微生物自身生理特性有关。但是无论添加CH3F还是NH4Cl,氢营养型产甲烷古菌丰度的增加意味着产甲烷途径由乙酸裂解产甲烷向乙酸互营氧化产甲烷迁移。这表明中温和高温石油烃降解产甲烷菌系中存在乙酸互营氧化产甲烷代谢过程,但是究竟是哪些乙酸互营氧化细菌和古菌参与了石油烃降解产甲烷过程,其对甲烷产生的贡献率,还有待于进一步的研究,目前我们正在应用同位素示踪结合微生物分子生态学技术揭示这些关键的细菌微生物类群。
| [1] | Ferry J G. Biochemistry of methanogenesis. Critical Reviews in Biochemistry and Molecular Biology, 1992, 27(6): 473-503. |
| [2] | Yilmaz V, Ince-Yilmaz E, Yilmazel YD. Is aceticlastic methanogen composition in full-scale anaerobic processes related to acetate utilization capacity? Applied Microbiology and Biotechnology, 2014, 98(11): 5217-5226. |
| [3] | Dolfing J. Thermodynamic constraints on syntrophic acetate oxidation. Applied and Environmental Microbiology, 2014, 80(4): 1539-1541. |
| [4] | Schnürer A, Schink B, Svensson BH. Clostridium ultunense sp. nov., a mesophilic bacterium oxidizing acetate in syntrophic association with a hydrogenotrophic methanogenic bacterium. International Journal of Systematic Bacteriology, 1996, 46(4): 1145-1152. |
| [5] | Westerholm M, Roos S, Schnürer A. Syntrophaceticus schinkii gen. nov., sp. nov., an anaerobic, syntrophic acetate‐oxidizing bacterium isolated from a mesophilic anaerobic filter. FEMS Microbiology Letters, 2010, 309(1): 100-104. |
| [6] | Westerholm M, Roos S, Schnürer A. Tepidanaero bacteracetatoxydans sp. nov., an anaerobic, syntrophic acetate-oxidizing bacterium isolated from two ammonium-enriched mesophilic methanogenic processes. Systematic and Applied Microbiology, 2011, 34(4): 260-266. |
| [7] | Hattori S, Kamagata Y, Hanada S. Thermacetogenium phaeum gen. nov., sp. nov., a strictly anaerobic, thermophilic, syntrophic acetate-oxidizing bacterium. International Journal of Systematic and Evolutionary Microbiology, 2000, 50(4): 1601-1609. |
| [8] | Balk M, Weijma J, Stams AJM. Thermotogalettingae sp. nov., a novel thermophilic, methanol-degrading bacterium isolated from a thermophilic anaerobic reactor. International Journal of Systematic and Evolutionary Microbiology, 2002, 52(4): 1361-1368. |
| [9] | Lee MJ, Zinder SH. Isolation and characterization of a thermophilic bacterium which oxidizes acetate in syntrophic association with a methanogen and which grows acetogenically on H2-CO2. Applied and Environmental Microbiology, 1988, 54(1): 124-129. |
| [10] | Hao LP, Lü F, He PJ. Predominant contribution of syntrophic acetate oxidation to thermophilic methane formation at high acetate concentrations. Environmental Science & Technology, 2010, 45(2): 508-513. |
| [11] | Schnürer A, Zellner G, Svensson BH. Mesophilic syntrophic acetate oxidation during methane formation in biogas reactors. FEMS Microbiology Ecology, 1999, 29(3): 249-261. |
| [12] | Hattori S. Syntrophic acetate-oxidizing microbes in methanogenic environments. Microbes and Environments, 2008, 23(2): 118-127. |
| [13] | Zengler K, Richnow HH, Rosselló-Mora R. Methane formation from long-chain alkanes by anaerobic microorganisms. Nature, 1999, 401(6750): 266-269. |
| [14] | Lovley DR. Anaerobes to the rescue. Science, 2001, 293(5534): 1444-1446. |
| [15] | Dolfing J, Larter SR, Head IM. Thermodynamic constraints on methanogenic crude oil biodegradation. The ISME Journal, 2007, 2(4): 442-452. |
| [16] | Garrity GM, Holt JG. Phylum AII. Euryarchaeotaphy. Nov//Bergey’s Manual of Systematic Bacteriology. 9th eds. New York: Springer, 2001: 211-355. |
| [17] | Townsend GT, Prince RC, Suflita JM. Anaerobic oxidation of crude oil hydrocarbons by the resident microorganisms of a contaminated anoxic aquifer. Environmental Science & Technology, 2003, 37(22): 5213-5218. |
| [18] | Gieg LM, Duncan KE, Suflita JM. Bioenergy production via microbial conversion of residual oil to natural gas. Applied and Environmental Microbiology, 2008, 74(10): 3022-3029. |
| [19] | Gray ND, Sherry A, Grant RJ. The quantitative significance of Syntrophaceae and syntrophic partnerships in methanogenic degradation of crude oil alkanes. Environmental Microbiology, 2011, 13(11): 2957-2975. |
| [20] | Siddique T, Penner T, Semple K. Anaerobic biodegradation of longer-chain n-alkanes coupled to methane production in oil sands tailings. Environmental Science & Technology, 2011, 45(13): 5892-5899. |
| [21] | Wang LY, Gao CX, Mbadinga SM. Characterization of an alkane-degrading methanogenic enrichment culture from production water of an oil reservoir after 274 days of incubation. International Biodeterioration & Biodegradation, 2011, 65(3): 444-450. |
| [22] | Cheng L, Ding C, Li Q. DNA-SIP reveals that Syntrophaceae play an important role in methanogenic hexadecane degradation. PLoS One, 2013, 8(7): e66784. |
| [23] | Jones DM, Head IM, Gray ND. Crude-oil biodegradation via methanogenesis in subsurface petroleum reservoirs. Nature, 2007, 451(7175): 176-180. |
| [24] | Gieg LM, Davidova IA, Duncan KE. Methanogenesis, sulfate reduction and crude oil biodegradation in hot Alaskan oilfields. Environmental Eicrobiology, 2010, 12(11): 3074-3086. |
| [25] | Mbadinga SM, Li KP, Zhou L. Analysis of alkane-dependent methanogenic community derived from production water of a high-temperature petroleum reservoir. Applied Microbiology and Biotechnology, 2012, 96(2): 531-542. |
| [26] | Cheng L, Rui J, Li Q. Enrichment and dynamics of novel syntrophs in a methanogenic hexadecane‐degrading culture from a Chinese oilfield. FEMS Microbiology Ecology, 2013, 83(3): 757-766. |
| [27] | Cheng L, He Q, Ding C. Novel bacterial groups dominate in a thermophilic methanogenic hexadecane‐degrading consortium. FEMS Microbiology Ecology, 2013, 85(3): 568-577. |
| [28] | Schwarz JIK, Lueders T, Eckert W. Identification of acetate-utilizing Bacteria and Archaea in methanogenicprofundal sediments of Lake Kinneret (Israel) by stable isotope probing of rRNA. Environmental Microbiology, 2007, 9(1): 223-237. |
| [29] | Conrad R, Klose M, Claus P. Pathway of CH4 formation in anoxic rice field soil and rice roots determined by13 C-stable isotope fractionation. Chemosphere, 2002, 47(8): 797-806. |
| [30] | Schnurer A, Nordberg A. Ammonia, a selective agent for methane production by syntrophic acetate oxidation at mesophilic temperature. Water Science & Technology, 2008, 57: 735-740. |
| [31] | Janssen PH, Frenzel P. Inhibition of methanogenesis by methyl fluoride: studies of pure and defined mixed cultures of anaerobic bacteria and archaea. Applied and Environmental Microbiology, 1997, 63(11): 4552-4557. |
| [32] | Macy JM, Snellen JE, Hungate RE. Use of syringe methods for anaerobiosis. The American Journal of Clinical Nutrition, 1972, 25(12): 1318-1323. |
| [33] | Ma T, Cheng L, Zheng Z, Qin Q, Dai L, Zhang H.Effects of pH on methanogenesis and methanogenic community in the cultures amended with acetate. Acta Microbiologica Sinica, 2014,54(12):1453 -1461.(in Chinese).麻婷婷,承磊,郑珍珍,覃千山,代莉蓉,张辉.不同pH缓冲液对由乙酸产甲烷菌群结构的影响.微生物学报,2014,54(12):1453-1461. |
| [34] | Ma T, Cheng L, Zheng Z, Qin Q, Dai L, Zhang H.The difference among different methanogenic hexadecane-degrading consortium’s DNA extraction method. China Biogas, 2014, 32(3): 3-7. (in Chinese).麻婷婷,承磊,郑珍珍,覃千山,张辉.石油烃降解产甲烷菌系DNA提取方法比较研究.中国沼气, 2014, 32(3): 3-7. |
| [35] | DeSantis TZ, Hugenholtz P, Larsen N. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Applied and Environmental Microbiology, 2006, 72(7): 5069-5072. |
| [36] | Schloss PD, Westcott SL, Ryabin T. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Applied and Environmental Microbiology, 2009, 75(23): 7537-7541. |
| [37] | Cole JR, Wang Q, Cardenas E. The ribosomal database project: improved alignments and new tools for rRNA analysis. Nucleic Acids Research, 2009, 37(suppl 1): D141-D145. |
| [38] | Good IJ. The population frequencies of species and the estimation of population parameters. Biometrika, 1953, 40(3-4): 237-264. |
| [39] | Boone DR. Strain GP6 is proposed as the neotypestrain of MethanothrixsoehngeniiVP pro synon. MethanothrixconciliiVP and MethanosaetaconciliiVPrequest for an opinion. International Journal of Systematic Bacteriology, 1991, 41(4): 588-589. |
| [40] | Patel GB, Sprott GD. Methanosaetaconcilii gen. nov., sp. nov.(“Methanothrixconcilii”) and Methanos aetathermoacetophila nom. rev., comb. nov.. International Journal of Systematic Bacteriology, 1990, 40(1): 79-82. |
| [41] | Mah RA. Isolation and characterization of Methanococcusmazei. Current Microbiology, 1980, 3(6): 321-326. |
| [42] | Barker HA. Studies upon the methane-producing bacteria. Archives of Microbiology, 1936, 7(1): 420-438. |
| [43] | Maestrojuan GM, Boone JE, Mah RA. Taxonomy and halotolerance of mesophilic Methanosarcina strains, assignment of strains to species, and synonymy of Methanosarcinamazei and Methanosarcinafrisia. International Journal of Systematic Bacteriology, 1992, 42(4): 561-567. |
| [44] | Mah RA, Kuhn DA. Transfer of the type species of the genus Methanococcus to the genus Methanosarcina, naming it Methanos arcinamazei (Barker 1936) comb. nov. et emend. and conservation of the genus Methanococcus (Approved Lists 1980) with Methanococcusvannielii (Approved Lists 1980) as the type species request for an opinion. International Journal of Systematic Bacteriology, 1984, 34(2): 263-265. |
| [45] | Wagner D, Schirmack J, Ganzert L. Methanos arcinasoligelidi sp. nov., a desiccation-and freeze-thaw-resistant methanogenic archaeon from a siberian permafrost-affected soil. International Journal of Systematic and Evolutionary Microbiology, 2013, 63(Pt 8): 2986-2991. |
| [46] | Liu Y, Boone DR, Sleat R. Methanosarcinamazei LYC, a new methanogenic isolate which produces a disaggregating enzyme. Applied and Environmental Microbiology, 1985, 49(3): 608-613. |
| [47] | Ma K, Liu X, Dong X. Methanos aetaharundinacea sp. nov., a novel acetate-scavenging methanogen isolated from a UASB reactor. International Journal of Systematic and Evolutionary Microbiology, 2006, 56(1): 127-131. |
| [48] | Cheng L, Qiu TL, Li X. Isolation and characterization of Methanoculleus receptaculi sp. nov. from Shengli Oil Field, China. FEMS Microbiology Letters, 2008, 285(1): 65-71.. |
| [49] | Cheng L, Dai L, Li X. Isolation and characterization of Methanother mobactercrinale sp. nov., a novel hydrogenotrophic methanogen from the Shengli Oil Field. Applied and Environmental Microbiology, 2011, 77(15): 5212-5219. |
| [50] | Zinder SH, Sowers KR, Ferry JG. Notes: Methanos arcinathermophila sp. nov., a thermophilic, acetotrophic, methane-producing bacterium. International Journal of Systematic Bacteriology, 1985, 35(4): 522-523. |
| [51] | Schink B. Energetics of syntrophic cooperation in methanogenic degradation. Microbiology and Molecular Biology Reviews, 1997, 61(2): 262-280. |
| [52] | Liu WT, Marsh TL, Cheng H. Characterization of microbial diversity by determining terminal restriction fragment length polymorphisms of genes encoding 16S rRNA. Applied and Environmental Microbiology, 1997, 63(11): 4516-4522. |
| [53] | Conrad R, Klose M. How specific is the inhibition by methyl fluoride of acetoclastic methanogenesis in anoxic rice field soil? FEMS Microbiology Ecology, 1999, 30(1): 47-56. |
| [54] | Yenigün O, Demirel B. Ammonia inhibition in anaerobic digestion: a review. Process Biochemistry, 2013, 48(5): 901-911. |
| [55] | Frenzel P, Bosse U. Methyl fluoride, an inhibitor of methane oxidation and methane production. FEMS Microbiology Ecology, 1996, 21(1): 25-36. |
| [56] | Conrad R, Klose M. Selective inhibition of reactions involved in methanogenesis and fatty acid production on rice roots. FEMS Microbiology Ecology, 2000, 34(1): 27-34. |
| [57] | Penning H, Conrad R. Effect of inhibition of acetoclastic methanogenesis on growth of archaeal populations in an anoxic model environment. Applied and Environmental Microbiology, 2006, 72(1): 178-184. |
| [58] | Hajarnis SR, Ranade DR. Revival of ammonia inhibited cultures of Methanobacterium bryantii and Methanos arcinabarkeri. Journal of Fermentation and Bioengineering, 1993, 76(1): 70-72. |
| [59] | Sprott GD, Patel GB. Ammonia toxicity in pure cultures of methanogenic bacteria. Systematic and Applied Microbiology, 1986, 7(2): 358-363. |
| [60] | Hunik JH, Hamelers HVM, Koster IW. Growth-rate inhibition of acetoclastic methanogens by ammonia and pH in poultry manure digestion. Biological Wastes, 1990, 32(4): 285-297. |
| [61] | Kadam PC, Boone DR. Influence of pH on ammonia accumulation and toxicity in halophilic, methylotrophic methanogens. Applied and Environmental Microbiology, 1996, 62(12): 4486-4492. |
| [62] | Chen Y, Cheng JJ, Creamer KS. Inhibition of anaerobic digestion process: a review. Bioresource Technology, 2008, 99(10): 4044-4064. |
| [63] | Sprott GD, Shaw KM, Jarrell KF. Ammonia/potassium exchange in methanogenic bacteria. Journal of Biological Chemistry, 1984, 259(20): 12602-12608. |
| [64] | Kato S, Sasaki K, Watanabe K. Physiological and transcriptomic analyses of the thermophilic, aceticlastic methanogen Methanos aetathermophil are sponding to ammonia stress. Microbes and Environments, 2014, 29(2): 162. |
| [65] | Zhang C, Yuan Q, Lu Y. Inhibitory effects of ammonia on methanogen mcrA transcripts in anaerobic digester sludge. FEMS Microbiology Ecology, 2014, 87(2): 368-377. |
| [66] | Fotidis I A, Karakashev D, Kotsopoulos TA. Effect of ammonium and acetate on methanogenic pathway and methanogenic community composition. FEMS Microbiology Ecology, 2013, 83(1): 38-48. |
| [67] | Steinhaus B, Garcia ML, Shen AQ. A portable anaerobic microbioreactor reveals optimum growth conditions for the methanogen Methanos aetaconcilii. Applied and Environmental Microbiology, 2007, 73(5): 1653-1658. |
| [68] | Westerholm M, Dolfing J, Sherry A. Quantification of syntrophic acetate‐oxidizing microbial communities in biogas processes. Environmental Microbiology Reports, 2011, 3(4): 500-505. |
| [69] | Nettmann E, Bergmann I, Pramschüfer S. Polyphasic analyses of methanogenic archaeal communities in agricultural biogas plants. Applied and Environmental Microbiology, 2010, 76(8): 2540-2548. |
| [70] | Fotidis IA, Karakashev D, Kotsopoulos TA. Effect of ammonium and acetate on methanogenic pathway and methanogenic community composition. FEMS Microbiology Ecology, 2013, 83(1): 38-48. |
 2015, Vol. 55
2015, Vol. 55