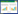中国科学院微生物研究所,中国微生物学会,中国菌物学会
文章信息
- 王璐, 杜德尧, 李金娥, 田宇清, 刘浩, 牛国清, 谭华荣. 2015
- Wang Lu, Du Deyao, Li Jin′e, Tian Yuqing, Liu Hao, Niu Guoqing, Tan Huarong. 2015
- 尼可霉素生物合成基因簇的改造及其异源表达
- Engineering and heterologous expression of a nikkomycin biosynthetic gene cluster
- 微生物学报, 2015, 55(6): 707-718
- Acta Microbiologica Sinica, 2015, 55(6): 707-718
-
文章历史
- 收稿日期:2014-12-19
- 修回日期:2015-01-19
2. 中国科学院微生物研究所,微生物资源前期开发国家重点实验室,北京 100101
2. State Key Laboratory of Microbial Resources, Institute of Microbiology, Chinese Academy of Sciences, Beijing 100101, China
尼可霉素(Nikkomycins)属于核苷肽类抗生素,主要产生菌有唐德链霉菌(Streptomyces tendae)[1]和圈卷产色链霉菌(Streptomyces ansochromogenes)[2]。尼可霉素的结构与几丁质合成酶的天然底物UDP-N-乙酰葡萄糖胺类似,是几丁质合成酶的强竞争性抑制剂,可以有效抑制丝状真菌、昆虫、蚜虫和酵母的生长[3]。尼可霉素由肽基和核苷两部分组成,肽基部分为羟基吡啶同型苏氨酸(hydroxypyridylhomothreonine,HPHT);核苷部分为尼可霉素C,在尼可霉素X和Z中(图1-A),同α-氨基己糖醛酸相连的碱基分别为4-甲酰-4-咪唑-2-酮和尿嘧啶,相应的核苷称为尼可霉素Cx和Cz。尼可霉素C与HPHT通过肽键相连形成二肽的尼可霉素X和Z,尼可霉素X和Z中的α-氨基已糖醛酸的6′-羧基分别与谷氨酸形成肽键,则形成三肽的尼可霉素I和J[4]。从唐德链霉菌和圈卷产色链霉菌中,分别克隆了尼可霉素生物合成基因簇,该基因簇包括一个调控基因和分布在三个转录单元中的21个结构基因(图1-B)。通过深入研究这些基因的功能,阐明了调控基因sanG通过调控sanO-sanV和sanN-sanI的转录,正调控尼可霉素的生物合成,而对sanF-sanX的转录没有影响[5, 6]。此外,阐明了尼可霉素的核苷和肽基的生物合成途径[7],推测两部分在合成酶SanS的作用下缩合形成有活性的尼可霉素[8],但相关研究至今未见报道。
|
| 图 1 尼可霉素的化学结构(A)及其生物合成基因簇(B) Fig.1 Chemical structures of nikkomycins (A) and organization of the gene cluster for nikkomycin biosynthesis (B). |
抗生素生物合成基因簇的异源表达,是研究抗生素生物合成和基因簇遗传改造的重要手段之一,其优势主要表现在:(1)将基因簇导入异源宿主中,通过检测终产物验证基因簇是否完整,通过构建系列突变株确定基因簇的边界[9];(2)异源表达是隐性基因簇激活的一个重要手段[10, 11];(3)异源表达可以提高目的产物的产量[12],而且由于异源宿主中特定修饰酶的存在或缺失,可以获得结构多样的目的产物[4];(4)与原始产生菌相比,基因簇在异源宿主中,更易于遗传操作[13];(5)不同的抗生素生物合成基因簇,在异源宿主中通过组合生物合成,形成新的杂合抗生素。
本研究将含有尼可霉素生物合成基因簇的重组质粒pNIK及其衍生质粒pNIKm(sanG和sanF的启动子分别替换为hrdB启动子),分别导入天蓝色链霉菌M1146中,获得菌株M1146-NIK和M1146-NIKm。通过对M1146-NIK和M1146-NIKm抗真菌活性的比较,评估了启动子替换对尼可霉素异源表达的影响;通过对有效活性组分的分离、鉴定,发现M1146-NIK与M1146-NIKm除了产生尼可霉素X和Z外,还可以产生尼可霉素Z的结构类似物—假尼可霉素Z。此外,我们比较了两株菌尼可霉素生物合成中间产物的产生情况。本研究将为尼可霉素核苷和肽基缩合机制的研究以及尼可霉素与其它核苷类抗生素的组合生物合成奠定基础。
1 材料和方法 1.1 材料 1.1.1 菌株:本文所用的菌株见表1。| Strains | Genotype/description | Reference/source |
| Escherichia coli | ||
| Top10 | F- mcrA Δ( mrr-hsdRMS-mcrBC) f80 lacZ ΔM15 Δ lacX74 recA1 araD139 Δ( ara leu) 7697 galU galK rpsL (StrR) endA1 nupG | Invitrogen |
| BW25113 | K-12 derivative, Δ araBAD, Δ rhaBAD | [ 14] |
| ET12567(pUZ8002) | dam dcm hsdS cat tet/pUZ8002 | [ 15] |
| Streptomyces | ||
| Streptomyces ansochromogenes 7100 (CGMCC 4. 321) | Wild-type, nikkomycin producer | China General Microbiological Culture Collection (CGMCC) |
| Streptomyces coelicolor M1146 | Δact Δred Δcpk Δcda | [ 13] |
| M1146-152 | S. coelicolor M1146 containing pSET152 | This work |
| M1146-NIK | S. coelicolor M1146 containing pNIK | This work |
| M1146-NIKm | S. coelicolor M1146 containing pNIKm | This work |
| Alternaria longipes | Indicator strain | CGMCC |
| Plasmids | Relevant characteristics | Reference/source |
| pBluescript II KS(+) | Routine cloning and subcloning vector | Stratagene |
| pSET152 | Integrative vector | [ 16] |
| pIJ790 | λ-RED (gam, bet, exo), cat, araC, rep101 ts | [ 14] |
| pIJ778 | aadA | [ 14] |
| pNIK | A pSET152 based plasmid containing the whole nikkomycin biosynthetic gene cluster | [ 17] |
| pBS-NIK | A pBluescript II KS(+) based plasmid containing the nikkomycin biosynthetic gene cluster | This work |
| pBS-hF | A pBluescript II KS(+) based plasmid containing part of sanF with hrdB promoter | This work |
| pBS-HI | A pBluescript II KS(+) based plasmid containing part of sanH, sanI and aadA | This work |
| pBS-HIhF | A pBluescript II KS(+) based plasmid containing part of sanF with hrdB promoter, part of sanH, sanI and aadA | This work |
| pSET152∷P hrdBG | pSET152 containing sanG with hrdB promoter | [ 18] |
| pBS-hG | pBluescript II KS(+) containing sanG with hrdB promoter | This work |
| pBS-hGX | pBluescript II KS(+) containing sanG with hrdB promoter, sanG downstream fragment and sanX fragment | This work |
| pSET152-hGX | pSET152 containing sanG with hrdB promoter, sanG downstream fragment and sanX fragment | This work |
| pBS-NIKaadA | A pBluescript II KS(+) based plasmid containing the whole nikkomycin biosynthetic gene cluster with PhrdB-driven sanF | This work |
| pBS-NIKm | A pBluescript II KS(+) based plasmid containing the whole nikkomycin biosynthetic gene cluster with PhrdB-driven sanG and sanF | This work |
| pNIKm | A pSET152 based plasmid containing the whole nikkomycin biosynthetic gene cluster with PhrdB-driven sanG and sanF | This work |
| Primers | Sequences (5′→3′) | Purpose |
| Underlined means restriction enzyme recognition sites. | ||
| sanH RT-F | GGCCTGGCTGGTCCTCAA | sanH RT-PCR |
| sanH RT-R | CTCGCCCCGCTCCTTCTC | sanH RT-PCR |
| sanHI-F | AATGAGCTCCACCCGGACCGGCTCGACTT | PCR 465 bp of sanH and sanI |
| sanHI-R | AATGCGGCCGCGTCGTCTCGGACGGCGGCTT | PCR 465 bp of sanH and sanI |
| sanF-F | GTGGTGCTGACGCTCGACTCGG | PCR 355 bp of sanF |
| sanF-R | CCGCTCGAGCCGTTGCGGTGGGTGTAGAGGC | PCR 355 bp of sanF |
| aadA-F | AATGCGGCCGCTGTAGGCTGGAGCTGCTTC | PCR of aadA cassett |
| aadA-R | AATGAATTCATTCCGGGGATCCGTCGACC | PCR of aadA cassett |
| hrdBp-F | AATTGAATTCCCGCCTTCCGCCGGAACG | PCR of hrdB promoter |
| hrdBp-R | GAACAACCTCTCGGAACGTTGA | PCR of hrdB promoter |
| sanG dn-F | AATCTCGAGCGGCCGGATCGTCGGTTCC | PCR 422 bp of sanG downstream region |
| sanG dn-R | AATGATATCCGGAAACCGCTGAAGCCCG | PCR 422 bp of sanG downstream region |
| sanX-F | AATGATATCGACCACGCTGCGCAACCA | PCR 147 bp of sanX |
| sanX-R | AATGGTACCTCAGGCCGGGTCCACACC | PCR 147 bp of sanX |
| sanG RT-F | GGCTTTCGGTGTGATGAC | sanG RT-PCR |
| sanG RT-R | CCTGGGAGACGAGGGTCT | sanG RT-PCR |
| sanF RT-F | GAAGCCCCGGTCTTCCCC | sanF RT-PCR |
| sanF RT-R | TGATCCGCTCGGTCCCCT | sanF RT-PCR |
| sanX RT-F | GCTCACGCCCTGCACCAT | sanX RT-PCR |
| sanX RT-R | GTCGACCCGCACCCCCAT | sanX RT-PCR |
| sanO RT-F | CGACGGCTGCCTGGAGTT | sanO RT-PCR |
| sanO RT-R | CGGATGCGGGCGATGAGA | sanO RT-PCR |
| sanI RT-F | GCACATCACCGCGGACAG | sanI RT-PCR |
| sanI RT-R | CAATCCTCGTCGATCCGG | sanI RT-PCR |
| sanN RT-F | CTGTCCGTGTCGGTCCTG | sanN RT-PCR |
| sanN RT-R | TCGATCTCGTGCGCTCTC | sanN RT-PCR |
| sanV RT-F | GGCGGAATGCTGCGAGAT | sanV RT-PCR |
| sanV RT-R | CGGGTGCGGGGATAAACG | sanV RT-PCR |
分子生物学常规操作参照分子克隆实验指南[19],链霉菌基因组的提取按照链霉菌操作手册进行[20]。
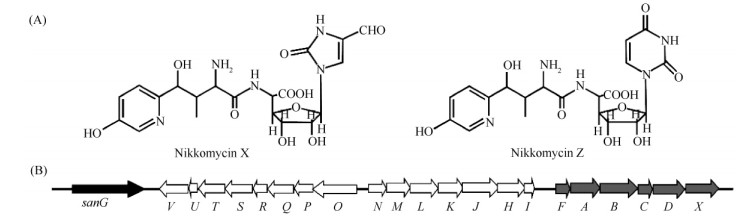
1.3 重组质粒的构建重组质粒pBS-HIhF和pSET152-hGX的构建流程如图2所示。具体步骤如下:以圈卷产色链霉菌基因组DNA为模板,用引物对sanF-F/sanF-R扩增sanF片段,用引物对sanHI-F/sanHI-R扩增sanHI片段;以天蓝色链霉菌基因组DNA为模板,用引物hrdBp-F和磷酸化的hrdBp-R扩增hrdB启动子片段;以pIJ778为模板,用引物对aadA-F/aadA-R扩增链霉素/壮观霉素抗性基因aadA片段。将sanF和hrdB启动子片段按照图2-A所示酶切,连入pBluescript II KS(+)得到重组质粒pBS-hF;将sanHI和aadA片段按照图2-A所示酶切,连入pBluescript II KS(+)得到重组质粒pBS-HI。用EcoR I/Xho I酶切pBS-hF,回收含有sanF和hrdB启动子的片段,连入相同酶切的pBS-HI,得到重组质粒pBS-HIhF。
|
| 图 2 重组质粒pBS-HIhF和pSET152-hGX的构建示意图 Fig.2 Schematic diagram of the construction of pBS-HIhF (A) and pSET152-hGX (B). Ph: hrdB promoter; aadA: Spectinomycin/streptomycin resistance gene; bla: Ampicillin resistance gene; aac(3)IV: Apramycin resistance gene; pSET152/EcoRV: EcoR V linearized pSET152. |
用Xba I/Xho I酶切pSET152∷hG,回收含有sanG及其上游hrdB启动子的片段,连入相同酶切的pBluescript II KS(+),得到重组质粒pBS-hG。以圈卷产色链霉菌基因组DNA为模板,用引物对sanG dn-F/SanG dn-R和sanX-F/sanX-R,分别扩增sanG下游片段Gdn和sanX片段。将两个扩增片段按照图2-B所示酶切,连入XhoI/KpnI双酶切的pBS-hG,得到重组质粒pBS-hGX。用EcoRV酶切pSET152,利用pSET152和pBluescript II KS(+)含有的lacZ同源序列,通过PCR targeting获得重组质粒pSET152-hGX。所有PCR扩增片段均由Invitrogen公司测序验证。
1.4 天蓝色链霉菌的接合转移重组质粒转化大肠杆菌 ET12567/pUZ8002,通过接合转移的方法[20]转入天蓝色链霉菌M1146。
1.5 尼可霉素的产生和分析尼可霉素的发酵和HPLC分析按照文献方法[17]进行,抗菌活性检测参照文献方法[21]。
1.6 RNA制备和RT-PCR分析RNA制备和cDNA合成按照文献[18]方法进行。PCR条件为95℃预变性5 min,28个循环:95℃变性45 s;60℃退火30 s;72℃延伸40 s,最后72℃延伸10 min。
1.7 尼可霉素的分离、纯化与鉴定发酵液通过大孔树脂(HP20),收集流出液;通过阳离子交换树脂,用0.2 mol/L氨水进行洗脱;将洗脱液通过葡聚糖凝胶柱Sephadex LH-20,分段收集并进行活性检测;将活性区段通过Agilent 1100 HPLC系统和ZORBAX SB-Aq柱(4.6 mm×250 mm,5 μm)进一步纯化;纯化样品的结构通过质谱与核磁共振波谱鉴定。
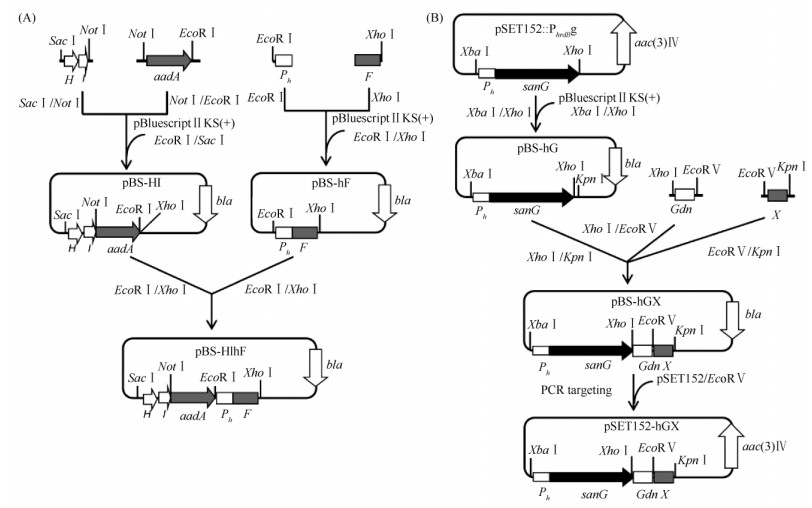
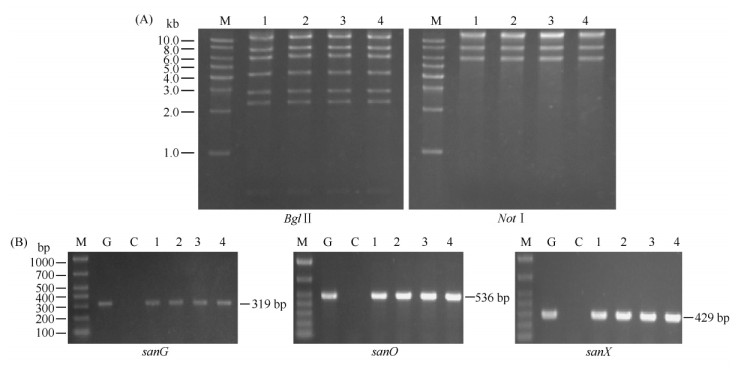
2 结果 2.1 重组质粒pNIKm的构建及验证鉴于sanG正调控sanO-sanV和sanN-sanI的转录,而对sanF-sanX的转录没有影响[5, 6]。为了尼可霉素生物合成基因簇的高表达,我们的策略是将sanG和sanF的启动子替换为组成型强启动子,需要通过3次PCR targeting和一次FLP重组酶介导的抗性基因切除来完成改造(图3)。首先,将pBluescript II KS(+)用EcoR V酶切,通过PCR targeting将尼可霉素生物合成基因簇从pSET152转移到pBluescript II KS(+);其次,以pBS-HIhF为模板,用引物sanHI-F和sanF-R,扩增HI-aadA-hF片段,通过PCR targeting将sanF的启动子替换为hrdB启动子;通过FLP重组酶的作用,将抗性基因aadA片段切除;最后,将pSET152-hGX用EcoR V酶切,通过PCR targeting将sanG的启动子替换为hrdB启动子,同时将pBluescript II KS(+)替换为pSET152,获得pNIKm(图3)。重组质粒pNIKm分别用Bgl II和Not I酶切验证。Bgl II酶切预期片段为12399、7609、6105、4108、2764、2259和534 bp;Not I酶切预期片段为21409、8062和6065 bp。重组质粒pNIKm酶切后琼脂糖凝胶电泳结果与预期一致(图4-A),说明构建的重组质粒pNIKm是正确的。
|
| 图 3 重组质粒pNIKm的构建示意图 Fig.3 Schematic diagram of the construction of pNIKm. pBluescript II KS(+)/EcoR V: EcoR V linearized pBluescript II KS(+); aadA: Spectinomycin/streptomycin resistance gene; Ph: hrdB promoter; pSET152/EcoR V: EcoR V linearized pSET152. |
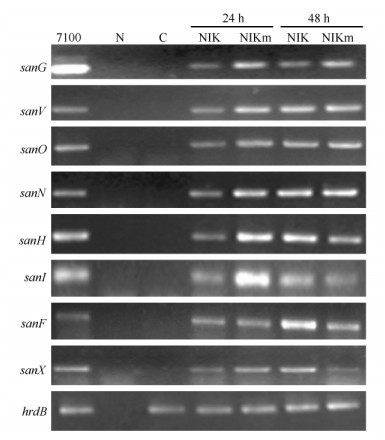
天蓝色链霉菌M1146是英国Mevyn Bibb教授课题组构建的异源表达菌株[13],目前该菌株已成功用于多种抗生素生物合成基因簇的表达[22]。分别将pNIK和pNIKm通过接合转移导入天蓝色链霉菌M1146中,得到M1146-NIK和M1146-NIKm。随机挑选四个接合子提取基因组DNA,选用三对引物(sanG RT-F/sanG RT-R、sanO RT-F/sanO RT-R和sanX RT-F/sanX RT-R)进行PCR验证。所有PCR扩增产物片段大小与预期一致(图4-B),说明pNIK和pNIKm成功转入天蓝色链霉菌M1146中。选取M1146-NIK和M1146-NIKm,培养24和48 h后分别提取RNA,以hrdB为对照,通过RT-PCR对尼可霉素生物合成基因簇中sanG、sanV、sanO、sanN、sanH、sanI、sanF和sanX的转录进行分析。结果表明:与对照菌株M1146-152相比,以M1146-NIK和M1146-NIKm的cDNA为模板,利用八对引物均能扩增出预期条带(图5),说明尼可霉素生物合成基因簇在天蓝色链霉菌M1146中成功转录。
|
| 图 4 重组质粒pNIKm的酶切验证以及M1146-NIK和M1146-NIKm的PCR验证 Fig.4 Confirmation of pNIKm and M1146-NIK/M1146-NIKm by restriction digestion analysis (A) and PCR amplifications (B). M: DNA ladder; 1, 2: Different clones of pNIK for restriction digestion or genomic DNAs from different M1146-pNIK as PCR templates; 3,4: Different clones of pNIKm for restriction digestion or genomic DNAs from different M1146-pNIKm as PCR templates; G: Genomic DNA from S. ansochromogenes 7100 as PCR template; C: Genomic DNA from M1146-152 as PCR template. |
|
| 图 5 尼可霉素生物合成基因簇的转录分析 Fig.5 RT-PCR analysis of genes involved in nikkomycin biosynthesis. PCR templates are genomic DNA from S. ansochromogenes 7100 (7100), ddH2O (N), genomic DNA from M1146-152 (C) and cDNA from M1146-NIK (NIK) or M1146-NIKm (NIKm). |
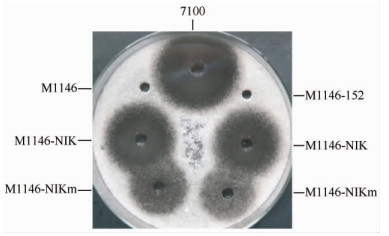
以上结果表明尼可霉素生物合成基因簇成功导入天蓝色链霉菌M1146中并正常转录,但这些基因能否合成有生物活性的尼可霉素分子?对尼可霉素生物合成基因簇的改造是否有助于尼可霉素在异源宿主中大量产生?为此,我们分别制备了M1146-NIK和M1146-NIKm的发酵液以及对照菌株M1146、M1146-152和圈卷产色链霉菌7100的发酵液,以长柄链格孢为指示菌,对其抗菌活性进行检测。结果表明:与M1146和M1146-152不同,M1146-NIK和M1146-NIKm对长柄链格孢有明显的抗菌活性;M1146-NIKm的抗菌活性低于M1146-NIK,且两者的抗菌活性均明显低于圈卷产色链霉菌7100 (图6)。
|
| 图 6 抗菌活性检测 Fig.6 Bioassays of nikkomycin production against A. longipes. |
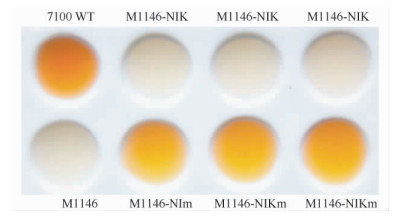
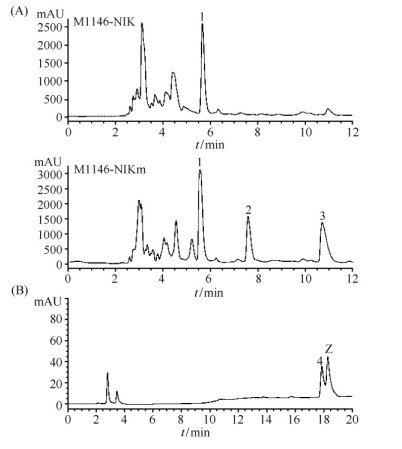
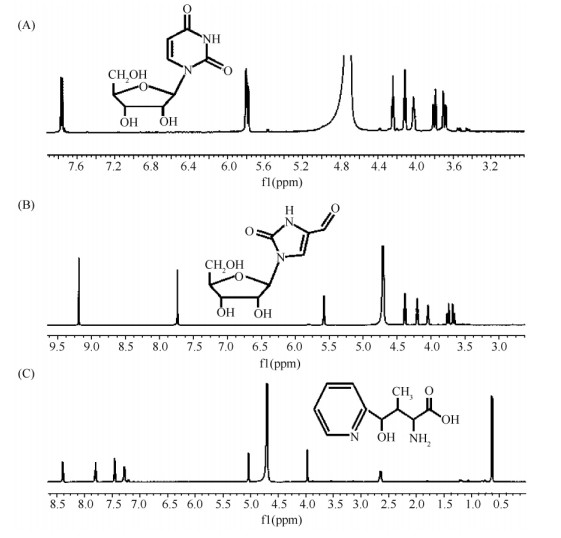
前期研究发现:巴比妥酸能够与尼可霉素核苷部分Cx的五元环碱基发生特异的颜色反应[23]。将M1146-NIK和M1146-NIKm的发酵液分别与巴比妥酸进行反应,结果发现:与圈卷产色链霉菌7100的发酵液一样,M1146-NIKm的发酵液能够与巴比妥酸反应显示橙色,而M1146-NIK的发酵液却不能显示橙色(图7)。进一步HPLC分析发现M1146-NIKm可以产生大量的化合物1、2和3,而M1146-NIK仅产生化合物1(图8-A),但是,直接对M1146-NIK和M1146-NIKm的发酵液进行HPLC分析,未检测到尼可霉素组分对应的吸收峰。通过化合物分离和核磁共振分析,化合物1、2和3结构鉴定为尿苷(uridine)、核糖基-4-甲酰-4-咪唑-2-酮(ribofuranosyl-4-formyl-4-imidazolone)和吡啶同型苏氨酸(pyridylhomothreonine,PHT)(图9、表4)。
|
| 图 7 巴比妥酸检测 Fig.7 Color reaction of compounds with barbituric acid. |
|
| 图 8 M1146-NIK和M1146-NIKm发酵液的HPLC分析(A)及假尼可霉素Z和尼可霉素Z的共进样HPLC分析(B) Fig.8 HPLC analysis of uridine (1), ribofuranosyl-4-formyl-4-imidazolone (2), PHT (3), nikkomycin pseudo-Z (4) and nikkomycin Z (Z). |
|
| 图 9 化合物1,2,3的结构鉴定 Fig.9 Structural determination of uridine, ribofuranosyl-4-formyl-4-imidazolone and PHT. A: Structure and 1H-NMR spectrum of uridine (1). B: Structure and 1H-NMR spectrum of ribofuranosyl-4-formyl-4-imidazolone (2). C: Structure and 1H-NMR spectrum of PHT (3). |
| Position | Uridine δ(1H, mult., J) | 13C (δ) | Ribofuranosyl-4-formyl-4-imidazolone | PHTδ(1H, mult., J) | 13C (δ) | |
| δ(1H, mult., J) | 13C (δ) | |||||
| 1 | 173.7 | |||||
| 2 | 151.9 | 153.2 | 3.97(1H, d, 4) | 58.8 | ||
| 3 | 2.65(1H, m) | 38.3 | ||||
| 4 | 166.4 | 124.2 | 5.03(1H, d, 5) | 76.0 | ||
| 5 | 5.79 (1H, d, 8.0) | 103.7 | 7.74 (1H, s) | 125.5 | 0.63(3H, d, 6.0) | 6.0 |
| 6 | 7.77 (1H, d, 8.0) | 143.6 | 9.19 (1H, s) | 180.2 | ||
| 1′ | 5.81 (1H, d, 4.0) | 91.5 | 5.57 (1H, d, 4.0) | 86.7 | ||
| 2′ | 4.24 (1H, t, 6.0 ) | 75.8 | 4.38 (1H, t, 6.0) | 73.4 | 159.8 | |
| 3′ | 4.11 (1H, t, 6.0) | 71.6 | 4.20 (1H, t, 6.0) | 70.0 | 7.45(1H, d, 8.0) | 123.2 |
| 4′ | 4.02 (1H, q, 4.0) | 83.6 | 4.04 (1H, q, , 4.0) | 84.5 | 7.80(1H, t, 8.0) | 138.3 |
| 5′ | 3.74 (2H, dd,8.0. 4.0) | 62.7 | 3.70 (2H, dd, 8.0, 4.0) | 60.9 | 7.28(1H, t, 8.0) | 120.9 |
| 6′ | 8.39(1H, d, 8.0) | 147.8 | ||||
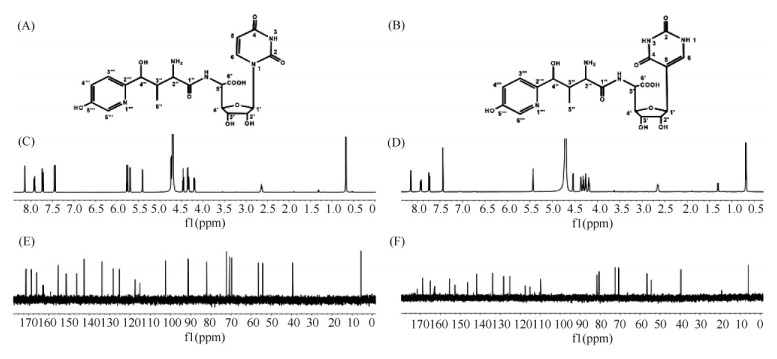
由于这3个化合物均不能抑制长柄链格孢的生长,为了纯化M1146-NIK和M1146-NIKm发酵液中的有效抗菌组分,分别对M1146-NIK和M1146-NIKm进行了发酵罐(5 L)培养,发酵液经过大孔树脂(HP 20)、离子交换树脂、Sephadex LH-20和HPLC等一系列样品浓缩和纯化,确定M1146-NIK可以产生尼可霉素X(约0.1 mg/L)和尼可霉素Z(约0.8 mg/L);同样M1146-NIKm也可以产生尼可霉素X(约0.1 mg/L)和尼可霉素Z(约0.2 mg/L)。在圈卷产色链霉菌7100中,尼可霉素X和Z的产量,分别为 220和120 mg/L[17]。此外,发现M1146-NIK与M1146-NIKm在尼可霉素X和Z之间还产生了一个新的吸收峰(化合物4),该化合物在M1146-NIK中产量约为0.4 mg/L,而在M1146-NIKm中产量约为0.06 mg/L。抗菌实验表明:与尼可霉素X和Z一样,化合物4对长柄链格孢有明显的抑制作用。化合物4与尼可霉素Z紫外吸收基本相同;高分辨质谱分析发现其分子量为496.1671,与尼可霉素Z的分子量496.1675相近;分子式为:C20H25N5O6,与尼可霉素Z的分子式相同,但共进样分析发现化合物4与尼可霉素Z不同(图8-B)。这些结果表明,化合物4与尼可霉素Z分子量相近、结构相似,可能为尼可霉素Z的结构类似物。将分离到的化合物4进行核磁共振波谱分析,与尼可霉素Z的1H-NMR和13C-NMR数据比较(图10、表5),发现化合物4与尼可霉素Z的化学位移信号相似,主要差异为:1H-NMR中尼可霉素Z的一个信号δ5.69(1H,d,J=4.5)在化合物4中向高场位移为δ4.71(1H,d,J=4.2),同时尼可霉素Z中δ5.75(1H,d,J =8.0)的信号峰在化合物4的1H-NMR中缺失;在13C-NMR中,尼可霉素Z的信号峰δ102.4和δ91.2在化合物4中分别变化为δ109.8和δ54.5。这些数据表明化合物4与尼可霉素Z的差别在核苷部分,且糖基部分与尿嘧啶可能连接在C-5上。尼可霉素Z中H-6(δ7.44,1H,d) 裂分由双峰变成单峰(δ7.44,1H,s)以及H-5与H-6在1H-1H COSY中无相关信号峰进一步验证了上述推测。结合耦合常数1H-1H COSY和1H-13C HSQC等其他数据以及文献报道[24],化合物4鉴定为假尼可霉素Z。
|
| 图 10 尼可霉素Z和假尼可霉素Z的结构鉴定 Fig.10 Structural determination of nikkomycin Z and pseudo-Z. Structure of nikkomycin Z (A) and pseudo-Z (B); 1H-NMR spectrum of nikkomycin Z (C) and pseudo-Z (D); 13C-NMR spectrum of nikkomycin Z (E) and pseudo-Z (F). |
| Position | Nikkomycin Z | Nikkomycin pseudo Z | ||
| δ( 1H, mult., J) | 13C (δ) | δ( 1H, mult., J) | 13C (δ) | |
| 2 | 151.5 | 152.7 | ||
| 4 | 166.1 | 165.1 | ||
| 5 | 5.75 (1H, d) | 102.4 | 109.8 | |
| 6 | 7.44 (1H, d) | 142.6 | 7.44 (1H, s) | 141.9 |
| 1′ | 5.69 (1H, d, 4.5) | 91.2 | 4.71 (1H, d, 4.0) | 54.5 |
| 2′ | 4.32 (1H, dd, 4.5, 5.5) | 72.2 | 4.18 (1H, dd, 4.0, 6.0) | 72.5 |
| 3′ | 4.46 (1H, t, 5.5) | 69.7 | 4.31 (1H, t, 6.0) | 70.7 |
| 4′ | 4.19 (1H, dd, 4.0, 5.5) | 82.1 | 4.25 (1H, dd, 4.0, 6.0) | 81.6 |
| 5′ | 4.74 (1H, d, 4.0) | 54.3 | 4.53 (1H, d, 4.0) | 80.5 |
| 6′ | 171.4 | 171.5 | ||
| 1″ | 168.7 | 168.9 | ||
| 2″ | 4.35 (1H, d, 4.0) | 56.4 | 4.36 (1H, d, 4.0) | 56.4 |
| 3″ | 2.64 (1H, m) | 39.6 | 2.65 (1H, m) | 39.7 |
| 4″ | 5.40 (1H, d) | 70.7 | 5.43 (1H, s) | 70.9 |
| 5″ | 0.67 (3H, d, 7.0) | 5.8 | 0.69 (3H, d, 7.0) | 5.9 |
| 2‴ | 8.70 (1H, s) | 146.3 | 8.68 (1H, s) | 146.4 |
| 3‴ | 7.92(1H, dd, 8.0, 6.0) | 125.2 | 7.92 (1H, dd, 8.0, 6.0) | 125.2 |
| 4′ | 7.72 (1H, dd, 8.0, 6.0) | 133.9 | 7.74 (1H, dd, 8.0, 6.0) | 143.9 |
| 5‴ | 155.5 | 155.4 | ||
| 6‴ | 8.14 (1H, d, 6.0) | 128.3 | 8.15 (1H, d, 6.0) | 128.3 |
在抗生素生物合成基因簇的异源表达中,为了基因簇在异源宿主中的成功表达,一个重要的手段就是将基因簇中关键基因的启动子替换为组成型启动子[25]。将谷氏菌素生物合成基因簇中关键结构基因的启动子替换为hrdB启动子,在异源宿主天蓝色链霉菌M1146中,谷氏菌素的产量提高了10倍[18]。在尼可霉素的生物合成中, sanG正调控sanO-sanV和sanN-sanI的转录,而对sanF-sanX的转录没有影响[5, 6]。本研究以含有尼可霉素生物合成基因簇的pNIK为出发质粒,将基因簇中sanG和sanF的启动子替换为hrdB启动子,构建了重组质粒pNIKm。将pNIK和pNIKm分别导入天蓝色链霉菌M1146中,获得菌株M1146-NIK和M1146-NIKm。虽然M1146-NIK和M1146-NIKm均有明显的抗真菌活性,但M1146-NIKm的抗菌活性明显低于M1146-NIK(图6)。发酵罐培养后,发现M1146-NIK和M1146-NIKm只能产生少量的尼可霉素X和Z。与M1146-NIK相比,M1146-NIKm除了大量积累尿苷外,还大量积累了核糖基-4-甲酰-4-咪唑-2-酮和PHT(图8-A),说明sanG和sanF的启动子替换为hrdB启动子后,尼可霉素基因簇中的大部分基因转录增强(图5),导致尼可霉素生物合成中间体大量积累。在尼可霉素的生物合成途径中,尿苷和核糖基-4-甲酰-4-咪唑-2-酮是尼可霉素核苷Cz和Cx的重要前体,而PHT是尼可霉素肽基HPHT的重要前体[7]。最近,有研究发现尿苷和核糖基-4-甲酰-4-咪唑-2-酮转化为Cz和Cx的过程中,需要烯醇式丙酮酸转移酶和氨基转移酶的参与[26, 27],但核苷部分的核糖如何经过酶促反应形成氨基核糖酸还需要深入研究。从现有结果来看,M1146-NIK和M1146-NIKm虽然大量积累了尼可霉素生物合成中间体,但尼可霉素X和Z的产量偏低,说明负责后续反应的基因在异源宿主中低水平表达或者其编码产物的酶活性较低。
将含有唐德链霉菌尼可霉素生物合成基因簇的两个质粒,同时导入变铅青链霉菌TK23中。在具有抗菌活性的6个转化子中,5个转化子的尼可霉素产量为10 mg/L,1个转化子的尼可霉素产量可以达到400-500 mg/L[28]。鉴于此,通过基因簇比较研究,筛选唐德链霉菌尼可霉素生物合成基因簇中可能参与核苷部分氨基核糖酸的基因,导入到M1146-NIKm中,通过检测尼可霉素的产生,确定这些基因是否参与核苷部分氨基核糖酸的转化。在此基础上,可以利用天蓝色链霉菌成熟的遗传操作平台,为解决尼可霉素核苷和肽基的缩合这一重要问题提供便利。
由于异源宿主与原始产生菌遗传背景的差异,异源宿主中特异性修饰酶的存在或缺失,产生结构多样化的目的产物[4]。本研究发现M1146-NIK和M1146-NIKm不仅产生了少量尼可霉素X和Z,同时也产生了假尼可霉素Z(图8-B和图10-B)。在圈卷产色链霉菌中,并未检测到假尼可霉素Z的存在,但在唐德链霉菌中已有假尼可霉素Z的报道[24]。假尼可霉素Z的前体为假尿嘧啶,但假尿嘧啶如何进入尼可霉素的生物合成途径?相应的酶如何识别假尿嘧啶和尿嘧啶,催化效率如何?对这些问题的回答,有待进一步的研究。综上所述,本文将对研究尼可霉素的生物合成,特别是氨基核糖酸的形成和核苷与肽基缩合机制的研究有促进作用,同时可以为尼可霉素与其它抗生素的组合生物合成奠定基础。
| [1] | Brillinger GU. Metabolic products of microorganisms. 181. Chitin synthase from fungi, a test model for substances with insecticidal properties. Archives of Microbiology, 1979, 121(1): 71-74. |
| [2] | Chen W, Zeng H, Tan H. Cloning, sequencing, and function of sanF: A gene involved in nikkomycin biosynthesis of Streptomyces ansochromogenes. Current Microbiology, 2000, 41(5): 312-316. |
| [3] | Dähn U, Hagenmaier H, Höhne H, König WA, Wolf G, Zähner H. Stoffwechselprodukte von mikroorganismen. 154. Mitteilung. Nikkomycin, ein neuer hemmstoff der chitinsynthese bei pilzen. Archives of Microbiology, 1976, 107(2): 143-160. |
| [4] | Niu G, Tan H. Nucleoside antibiotics: biosynthesis, regulation, and biotechnology. Trends in Microbiology, 2015, In press. |
| [5] | He X, Li R, Pan Y, Liu G, Tan H. SanG, a transcriptional activator, controls nikkomycin biosynthesis through binding to the sanN-sanO intergenic region in Streptomyces ansochromogenes. Microbiology, 2010, 156(Pt 3): 828-837. |
| [6] | Liu G, Tian Y, Yang H, Tan H. A pathway-specific transcriptional regulatory gene for nikkomycin biosynthesis in Streptomyces ansochromogenes that also influences colony development. Molecular Microbiology, 2005, 55(6): 1855-1866. |
| [7] | Winn M, Goss RJ, Kimura K, Bugg TD. Antimicrobial nucleoside antibiotics targeting cell wall assembly: recent advances in structure-function studies and nucleoside biosynthesis. Natural Product Reports, 2010, 27(2): 279-304. |
| [8] | Li J, Li L, Tian Y, Niu G, Tan H. Hybrid antibiotics with the nikkomycin nucleoside and polyoxin peptidyl moieties. Metabolic Engineering, 2011, 13(3): 336-344. |
| [9] | Niu G, Li L, Wei J, Tan H. Cloning, heterologous expression, and characterization of the gene cluster required for gougerotin biosynthesis. Chemistry & Biology, 2013, 20(1): 34-44. |
| [10] | Liu G, Chater KF, Chandra G, Niu G, Tan H. Molecular regulation of antibiotic biosynthesis in streptomyces. Microbiology and Molecular Biology Reviews, 2013, 77(1): 112-143. |
| [11] | Niu G, Tan H. Biosynthesis and regulation of secondary metabolites in microorganisms. Science China. Life Sciences, 2013, 56(7): 581-583. |
| [12] | Komatsu M, Uchiyama T, Omura S, Cane DE, Ikeda H. Genome-minimized Streptomyces host for the heterologous expression of secondary metabolism. Proceedings of the National Academy of Sciences of the United States of America, 2010, 107(6): 2646-2651. |
| [13] | Gomez-Escribano JP, Bibb MJ. Engineering Streptomyces coelicolor for heterologous expression of secondary metabolite gene clusters. Microbial Biotechnology, 2011, 4(2): 207-215. |
| [14] | Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proceedings of the National Academy of Sciences of the United States of America, 2000, 97(12): 6640-6645. |
| [15] | MacNeil DJ, Gewain KM, Ruby CL, Dezeny G, Gibbons PH, MacNeil T. Analysis of Streptomyces avermitilis genes required for avermectin biosynthesis utilizing a novel integration vector. Gene, 1992, 111(1): 61-68. |
| [16] | Bierman M, Logan R, O'Brien K, Seno ET, Rao RN, Schoner BE. Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene, 1992, 116(1): 43-49. |
| [17] | Liao G, Li J, Li L, Yang H, Tian Y, Tan H. Cloning, reassembling and integration of the entire nikkomycin biosynthetic gene cluster into Streptomyces ansochromogenes lead to an improved nikkomycin production. Microbial Cell Factories, 2010, 9: 6. |
| [18] | Du D, Zhu Y, Wei J, Tian Y, Niu G, Tan H. Improvement of gougerotin and nikkomycin production by engineering their biosynthetic gene clusters. Applied Microbiology and Biotechnology, 2013, 97(14): 6383-6396. |
| [19] | Sambrook P, Fritsch EF, Maniatis T. Molecular Cloning: a Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press, 2001. |
| [20] | Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA. Practical Streptomyces Genetics. Norwich, UK: John Innes Foundation, 2000. |
| [21] | Feng C, Ling H, Du D, Zhang J, Niu G, Tan H. Novel nikkomycin analogues generated by mutasynthesis in Streptomyces ansochromogenes. Microbial Cell Factories, 2014, 13(1): 59. |
| [22] | Gomez-Escribano JP, Bibb MJ. Heterologous expression of natural product biosynthetic gene clusters in Streptomyces coelicolor: from genome mining to manipulation of biosynthetic pathways. Journal of Industrial Microbiology & Biotechnology, 2014, 41(2): 425-431. |
| [23] | Delzer J, Fiedler HP, Müller H, Zähner H, Rathmann R, Ernst K, König WA. New nikkomycins by mutasynthesis and directed fermentation. The Journal of Antibiotics, 1984, 37(1): 80-82. |
| [24] | Heitsch H, König WA, Decker H, Bormann C, Fiedler HP, Zähner H. Metabolic products of microorganisms. 254. Structure of the new nikkomycins pseudo-Z and pseudo-J. The Journal of Antibiotics, 1989, 42(5): 711-717. |
| [25] | Stevens DC, Hari TP, Boddy CN. The role of transcription in heterologous expression of polyketides in bacterial hosts. Natural Product Reports, 2013, 30(11): 1391-1411. |
| [26] | Binter A, Oberdorfer G, Hofzumahaus S, Nerstheimer S, Altenbacher G, Gruber K, Macheroux P. Characterization of the PLP-dependent aminotransferase NikK from Streptomyces tendae and its putative role in nikkomycin biosynthesis. The FEBS Journal, 2011, 278(21): 4122-4135. |
| [27] | Oberdorfer G, Binter A, Ginj C, Macheroux P, Gruber K. Structural and functional characterization of NikO, an enolpyruvyl transferase essential in nikkomycin biosynthesis. The Journal of Biological Chemistry, 2012, 287(37): 31427-31436. |
| [28] | Bormann C, Möhrle V, Bruntner C. Cloning and heterologous expression of the entire set of structural genes for nikkomycin synthesis from Streptomyces tendae Tü901 in Streptomyces lividans. Journal of Bacteriology, 1996, 178(4): 1216-1218. |
 2015, Vol. 55
2015, Vol. 55