中国科学院微生物研究所,中国微生物学会,中国菌物学会
文章信息
- 宋晓丹, 张园, 邹祥. 2018
- Xiaodan Song, Yuan Zhang, Xiang Zou. 2018
- TOR信号调控真菌细胞的生长与代谢
- Fungal cell growth and metabolism regulated by the TOR signal pathway
- 微生物学报, 58(10): 1691-1700
- Acta Microbiologica Sinica, 58(10): 1691-1700
-
文章历史
- 收稿日期:2017-11-20
- 修回日期:2018-01-24
- 网络出版日期:2018-03-08
雷帕霉素靶(target of rapamycin,TOR)蛋白是真核细胞生长的关键调控因子,是一类进化上保守的丝氨酸/苏氨酸(Ser/Thr)蛋白激酶,属于磷脂酰肌醇相关激酶(phosphatidylinositol kinase-related kinases,PIKKs)家族。Rapamycin(Rap)是来源于吸水链霉菌(Streptomyces hygroscopicus)的一种大环内酯类免疫抑制剂,用于同种异体移植排斥反应的临床治疗[1]。在酿酒酵母(Saccharomyces cerevisiae)细胞作用的深入研究中发现Rap可结合并抑制FKBP (FK506-binding protein)蛋白,不可逆地抑制细胞周期的G1时期而控制细胞生长[2]。Michael等在具有Rap抗性的FKBP蛋白功能缺陷型酿酒酵母中发现TOR1或TOR2基因也被改变,而敲除TOR1或TOR2基因与Rap处理均抑制细胞生长,因此认为TOR1和TOR2是FKBP-Rap复合物的靶位,而FKBP-Rap复合物抑制TOR活性[3],并由此获得2017年拉斯克基础医学研究奖(2017 Albert Lasker Basic Medical Research Award)。自TOR蛋白在酵母细胞发现后,在其他真菌、果蝇、植物以及哺乳动物等真核生物中也相继被发现,且TOR信号通路功能保守,对自身生长和发展非常重要[4]。在植物、蠕虫、果蝇以及哺乳动物细胞等中有一个TOR编码基因,而酿酒酵母中存在两个TOR编码基因(70%同源性)[5]。真菌细胞中,TOR蛋白复合物多样性产生不同分支功能的信号通路,是细胞TOR作为生命活动调节的重要因素。因此,本文综述近几年真菌TOR信号途径对细胞生长和代谢的研究进展。
1 TOR的结构真菌细胞TOR蛋白是一类分子量较大的蛋白质(约280 kDa),所有TOR蛋白有相似的结构域(图 1)。TOR蛋白从N端到C端整齐地排列着HEAT重复序列、FAT、FRB、激酶以及FATC等结构域。整个结构域N端的一半是由2个HEAT重复基序组成。HEAT能够形成一对相互作用的反向平行α-螺旋,HEAT repeats是TOR复合物亚基结合的区域[6]。中心部位的FAT结构域、C端末尾的FATC结构域是PIKK的家族成员,2个结构域的互作可能参与调控激酶活性[7]。FRB结构域是FKBP-Rap复合物结合区域,所有TOR突变引起的Rap抗性,都是FRB结构域破坏引起[5]。Rap仅仅对TOR Complex 1 (TORC1)有抑制作用,Rap首先和细胞内的FKBP结合,再与TOR1/2的FRB结构域结合,从而抑制TOR激酶的活性[8]。

|
| 图 1 TOR结构域 Figure 1 The structure domain of the TOR in yeast. |
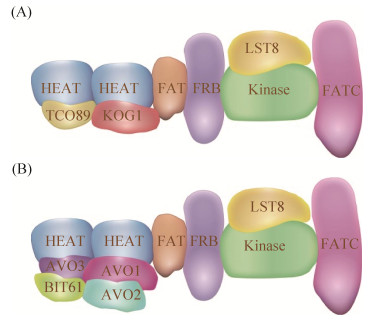
TOR能够和多种蛋白结合形成复合物如TORC1和TOR Complex 2 (TORC2),在酵母细胞TORC1中TOR1或TOR2能够和KOG1、TCO89、LST8结合形成TORC1 (图 2-A),其中KOG1与HEAT结合,有助于底物进入TOR激酶的催化区域[9],TCO89接收来自EGO复合物(Ego1、Ego2、Ego3)信号,而LST8与TOR激酶结合稳定激酶结构[5]。TOR2与AVO1、AVO2、AVO3、LST8以及BIT61 (与BIT2同源)形成复合物TORC2,其中AVO1和AVO3结合于TOR2的HEAT区域,加强TOR2的稳定性,LST8与TOR2激酶结合,稳定并促进激酶活性,AVO2和BIT61的功能还不清楚[10-12] (图 2-B)。TORC1能感知环境中的营养物质、生长因子、压力等信号,来参与调节蛋白质翻译、核糖体合成、基因转录、蛋白质降解以及自噬等信号通路。此外酵母细胞可通过固定在液泡/溶酶体膜上的EGO复合物与Gtr1/2相互作用激活TORC1活性,为亮氨酸或其他氨基酸依赖的Gtr激活机制[13],近期发现谷氨酰胺(Glutamine,Gln)也能够直接激活TORC1而非Gtr依赖型[14]。TORC1的调控通路中,主要存在两类调控分支:AGC激酶Sch9 (与哺乳动物细胞的S6K同源)支路和Tap42-Ppase复合物调控支路,分别调控细胞的生长和代谢[15]。
|
| 图 2 真菌酵母TORC1 (A)和TORC2 (B)结构 Figure 2 The structures of the TORC1 (A) and TORC2 (B) in yeast. |
2 TOR与细胞生长和自噬 2.1 TOR调控细胞生长
真菌细胞生长能够根据自身需求进行动态调控,其中核糖体生成(ribosome biogenesis,Ribi)起到非常重要作用。核糖体的生成是需要多种因子和RNAs共同协作的动态复杂过程。细胞根据不同生长环境调节核糖体各组分比例以保证蛋白正常合成,合成核糖体需要3个主要的RNA polymerases (Pols)对核糖体组分的精确协调,主要包括PolⅠ、PolⅡ以及Pol Ⅲ,其中PolⅠ主要负责合成核糖体RNA (ribosomal RNA,rRNA);PolⅡ合成核糖体蛋白(ribosomal protein,RP);Pol Ⅲ主要合成5S rRNA和转运RNA (transfer RNA,tRNA)[16]。
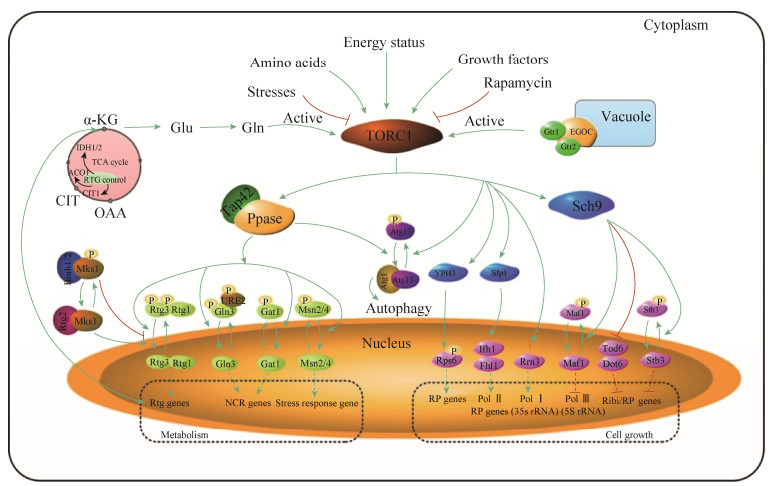
TORC1参与核糖体合成、转录起始以及进入G0阶段[17]需要通过Sch9介导,即Sch9是TORC1蛋白的直接底物,也有报道TORC1通过YPK3促进核糖体蛋白Rps6的磷酸化进一步促进核糖体蛋白的合成[18](图 3)。禾谷镰刀菌(Fusarium graminearum)中发现TOR可能通过Sch9调控细胞的高渗甘油响应、压力响应以及次级代谢产物合成,而敲除Sch9则影响细胞气生菌丝增长和分生孢子的形成,增加细胞对渗透压、氧化应激、Rap以及细胞壁损伤的药物敏感性;另外Sch9可与下游Maf1蛋白互作,调节霉菌毒素(脱氧雪腐镰刀菌烯醇)合成[19]。此外,烟曲霉菌(Aspergillus fumigatus)与酵母Sch9同源的SchA蛋白,也发现参与渗透压、氧化应激等压力应激响应[20]。
|
| 图 3 TORC1调控酵母细胞的生长与代谢 Figure 3 Cell growth and metabolism regulated by TORC1 in yeast. |
Ribi和RP转录抑制因子Dot6、Tod6及Stb3能促进RPD3L组蛋白去乙酰化酶复合物与Ribi和RP基因启动子结合而发挥转录抑制作用,即抑制Ribi和RP基因合成。Dot6和Tod6调控Ribi调节子影响细胞大小,而Stb3影响调控RP基因表达而影响细胞增长率。Stb3、Dot6和Tod6蛋白的磷酸化水平影响其抑制活性,TORC1-Sch9通路抑制Stb3、Dot6和Tod6磷酸化,而促进Ribi和RP的合成和表达[21](图 3)。转录因子Sfp1能够促进Fhl1和Ifh1蛋白与RP基因启动子结合而激活RP基因(图 3),影响Ribi/RP基因的表达效率,而Sfp1敲除使细胞生长缓慢、Ribi/RP基因表达抑制以及细胞体积变小[22-23]。
Pol Ⅲ的抑制因子Maf1,通过Rap处理或营养限制等环境信号去磷酸化并定位于细胞核对5S rRNA和tRNA产物进行剪切并抑制产物正常合成。当处于营养充足条件下,借助TORC1-Sch9通路磷酸化调节,使Maf1完成细胞核到细胞质的转换,增加Pol Ⅲ与转录基因结合,促进5S rRNA和tRNA基因的转录[24-25](图 3)。在理想生长状况下,TORC1能够促进Pol Ⅲ亚基Rpc82的类泛素化而促进Pol Ⅲ各亚基的组装从而激发tRNA的转录[26]。
此外,PolⅠ与Rrn3翻译起始因子结合促进了PolⅠ对rDNA的转录,而TORC1的细胞核定位能够促进35S rRNA的合成及细胞生长[27-28] (图 3),但Rrn3因子是否通过TORC1的磷酸化而调控PolⅠ对rDNA的转录缺乏文献报道,而TORC1对PolⅠ的调控机制仍然不清楚。
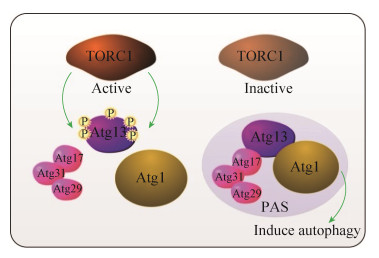
2.2 TOR调控细胞自噬真核细胞自噬是一种蛋白质降解过程,在饥饿状态下,细胞内的核糖体、蛋白质以及多余的细胞器通过自噬体转运到溶酶体和液泡,转化成为营养物质以适应长期的饥饿环境[16]。酵母细胞中的自噬相关基因(autophagy-related genes,Atg) Atg13是TORC1的直接底物,在营养物质充裕时,被过度磷酸化,营养限制或Rap处理时,Atg13去磷酸化,并加强了与自噬蛋白Atg17的相互作用,与Atg29、Atg31形成复合物,即Atg13-Atg17- Atg31-Atg29。一个自噬体的前体结构/吞噬泡聚集位点(phagophore assembly site,PAS)如图 3、4所示。Atg1激酶与Atg13结合时,Atg1蛋白激酶的活性增强。营养物质丰富时,Atg1激酶的活性最小,自噬抑制;当处于饥饿状态时,Atg1激酶活性增强,诱导自噬[29]。TORC1也可直接控制Atg1激酶而诱导自噬[30]。TOR与自噬存在双向调控,在氮限制条件下,TORC1在一定程度上可被自噬激活,而在长时间氮限制条件下,Atg13也可被Atg1激酶重新磷酸化[31]。在促进自噬方面,PP2A也发挥着非常重要的作用,当TORC1失活时,2个PP2A磷酸酶蛋白被激活(PP2A-Cdc55和PP2A-Rts1),并使Atg13充分去磷酸化,并诱导自噬(图 3)。而PP2A敲除,Rap处理后Atg13去磷酸化、Atg1激酶活化、PAS的形成以及自噬的诱导等机制都会造成损伤[32]。在盘基网柄菌(Dictyostelium discoideum)细胞中,Rap通过促进细胞体内的活性氧和胞内钙离子积累而诱导自噬产生[33]。酵母细胞生存环境中Zn的缺乏,能抑制TORC1活性,而诱导非选择性自噬产生[34]。酵母细胞胞外蛋白Ecm33能有效促进细胞吸收葡萄糖并激活TORC1信号通路,而Ecm33蛋白缺失时,即使细胞处于营养丰富条件下,也能通过影响Atg13和Sch9去磷酸化而诱导细胞自噬形成[35]。
|
| 图 4 酵母细胞TORC1调控细胞自噬 Figure 4 Cell autophagy regulated by TORC1 in yeast. |
3 TORC1与代谢
已有证据表明,TORC1介导下游许多信号通路调控细胞代谢过程,其中氮分解代谢阻遏(Nitrogen catabolite repression,NCR)效应最为常见。通过NCR通路调控细胞对氮源的吸收与利用,而NCR基因表达易受到胞内氨基酸浓度的影响。当真菌细胞处于氮充足状态,TORC1下游的PP2A分支保持低活性;在外界压力或氮饥饿状态时,细胞通过Sch9分支抑制蛋白质和核糖体合成;在不缺乏葡萄糖(能量)情况下,氮饥饿通过PP2A支路对氮源进行同化吸收以及促进氨基酸合成[15]。优势氮源铵盐(NH4+)、谷氨酰胺(Glutamine,Gln)及谷氨酸盐(Glutamate,Glu)是氨基酸合成的主要前体物质,而Gln和Glu的合成主要依赖于TCA循环α-酮戊二酸的合成能力[36],TORC1介导逆行响应(retrograde response,RTG)信号通路参与调控糖酵解和TCA循环合成α-酮戊二酸。细胞在压力胁迫环境中,TORC1积极调控胁迫应答转录因子Msn2/4进行响应,缓解压力环境带来的细胞损害。
3.1 TORC1与氮代谢真菌细胞能够感知环境中含氮营养物质,并根据其来调整自身的转录、代谢合成等生命过程。通常会利用Gln、Glu或者NH4+等优先利用型氮源,然后利用脯氨酸(Proline,Pro)、尿素以及尿囊素等非优先利用型氮源。调控NCR基因表达的转录因子主要有激活转录因子如Gln3和Gat1/Nil1/AreA及抑制转录因子Gzf3/Nil2/Deh1和Dal80/Uga43/AreB等结合于启动子的GATA序列[37]。TORC1主要通过Gat1和Gln3两个转录因子调控NCR基因,当细胞处在优先利用型氮源环境时,TORC1以及磷酸酶(Sit4或PP2A)被激活,Gln3和Gat1磷酸化并滞留在细胞质中,抑制NCR基因表达,而Gln3在胞质中定位需要与URE2形成复合物[38-39]。当细胞处于非优先型氮源环境,Gln3和Gat1去磷酸化,并定位于细胞核,促进NCR基因转录[40-41](图 3)。
光滑念珠菌(Candida glabrata)中Gln3是氮同化吸收主要的调控者[42]。在水稻恶苗病菌(Fusarium fujikuroi)中,AreA和AreB不仅仅调控真菌氮代谢,也参与调控碳代谢、转运以及次级代谢等过程,如赤霉素、红色比卡菌素、镰红菌素、伏马菌素等的合成[37]。S. cerevisiae通过Ehrlich通路合成的2-苯基乙醇,在Gat1和Gln3过表达后,能够大幅度增加2-苯基乙醇产量[40],而烟曲霉(A. fumigatus)中的TOR蛋白不仅能调控线粒体功能,还可调控鸟氨酸代谢。鸟氨酸代谢可促进铁载体的形成,而抑制TOR蛋白也伴随着铁离子调控功能的缺失[43]。
我们前期研究发现,类酵母真菌出芽短梗霉(Aureobasidium pullulans)的氮压力环境影响细胞生长和代谢产物聚苹果酸的合成。氮限制压力可促进聚苹果酸合成,进一步用Rap处理后,能明显抑制出芽短梗霉细胞生长及聚苹果酸的合成,并下调TOR1、聚苹果酸合成相关基因的表达水平。研究表明TOR信号通路参与聚苹果酸合成的代谢调控[44]。
近期,Liu等[45]首次发现磷酸是TOR信号通路调控的第三种营养物质。除了碳、氮外,细胞可以通过磷酸转运蛋白感知磷酸浓度变化来调控TOR蛋白激酶活性,调节致病菌白色念珠菌(Candida albicans)的生长和形态转换,以适应宿主环境变化得以存活。此外磷酸盐转运蛋白Pho84通过Gtr1间接激活TORC1,参与细胞内的调控磷酸盐稳态,对Pho84突变会使细胞对Rap非常敏感及TORC1失活,而Gtr1过表达以及持续激活Gtr1能够恢复TORC1活性。
3.2 TORC1与逆行响应调控线粒体具有自身可编码的DNA,但多数线粒体蛋白(> 99%)在核中编码,因而细胞核和线粒体之间的信息传递非常重要,特别在线粒体功能失调时与细胞核的信息传递并触发细胞核的补偿响应(线粒体→细胞核),即逆行响应(retrograde response,RTG),而机体线粒体功能障碍会激活细胞内的RTG信号通路[46]。酵母细胞对其他氮源的吸收不同于对谷氨酰胺和谷氨酸,是通过将其他氮源转变成铵盐,再通过糖酵解途径和TCA循环通路产生的α-酮戊二酸形成Gln或Glu。而RTG通路的意义在于线粒体功能障碍时或氮源贫瘠条件下,增强TCA循环活力对铵盐进行同化吸收[22]。RTG调控通路有正向调控因子Rtg1、Rtg2、Rtg3以及Grr1和负向调控因子Mks1、Bmh1、Bmh2以及LST8[47]。
同TORC1对Gat1和Gln3的调控类似,当线粒体的功能完整且环境氮源充足时,TORC1使Rtg3和Mks1高度磷酸化,Mks1与Bmh1/2形成复合物,将Rtg1/ 3异质二聚物保留在细胞质中;当线粒体功能紊乱且处于氮限制条件下,Rtg3去磷酸化,Rtg2与抑制因子Msk1结合,促进Rtg1/3进入细胞核并诱导靶基因表达[22](图 3)。Rtg1/3的定位也通过Rtg2感知细胞内部ATP变化,而ATP流变化又通过EGO复合物介导TORC1进行下游调节[48]。Rtg2蛋白有3个使ATP稳定结合的激活位点,且能够控制酿酒酵母的寿命[49]。
参与调控TCA循环的RTG靶基因主要调控TCA循环通路的前3个基因CIT (柠檬酸合成酶的线粒体同工酶)、ACO1 (顺乌头酸酶)以及IDH1/2 (NAD+依赖型异柠檬酸脱氢酶)(图 3),另外RTG调控的CIT2 (过氧化物酶体柠檬酸合酶)参与调控乙醛酸循环[50]。细胞中Rtg1/3蛋白调控TCA循环中的4个基因的表达,在线粒体功能障碍条件下确保合成代谢过程正常进行,如谷氨酸等。
RTG通路通过维持TCA循环代谢流调控乙醇依赖型的酵母丝状生长[51]。TORC1调控的PP2A支路下游的Gat1、Gln3及Rtg1/3转录因子,调控中性脂肪的合成,Gat1、Gln3的敲除能够破坏脂肪动态平衡[52]。酿酒酵母细胞中Rtg1/2敲除突变不仅降低了线粒体自噬还提高线粒体活性,这种突变削弱了细胞移除过氧化氢的能力,而RTG信号能够提高细胞对过氧化氢的抗逆性能力[53]。ADR1 (调控过氧化物酶合成、β-氧化及非发酵型碳源利用)和CAT8 (编码糖异生和乙醛酸循环酶)能够激活Rtg2,并在棉子糖条件下与其互作增强了醋酸诱导型细胞程序性死亡的抵抗能力[54]。
3.3 TORC1与胁迫响应细胞面对压力胁迫环境时需要调整胁迫响应基因转录水平,以快速适应胁迫环境。Msn2和Msn4是调控多数应激反应的2个锌指结构转录因子,可参与多种类型的胁迫响应,如氧化应激、热激、渗透应激及饥饿等胁迫。胁迫环境中,Msn2/4结合于胁迫应答元件(stress-responsive element,STRE)并激活大量的胁迫应答基因转录[55]。无压力胁迫时,TORC1促进Msn2/4磷酸化并定位于胞质;压力胁迫时,Msn2/4快速去磷酸化定位于细胞核,并结合于STRE的GGGGA区域,促进靶基因转录(图 3),如Msn2/4氧化应激响应中,H2O2诱导Msn2/4进入细胞核与Ctt1 (胞质过氧化氢酶)启动子结合,促进Ctt1表达,维持氧化还原平衡[56]。Msn2/4不仅对胁迫环境产生应激响应,还激活相应代谢通路。
糖类物质合成需要Ugp1 (UDP-G焦磷酸化酶)编码的糖基供体UDP-Glc (尿苷二磷酸-葡萄糖)参与,而胁迫应答转录因子Msn2/4直接与Ugp1启动子结合调控Ugp1表达,在各种压力胁迫状态下,Msn2/4都能够诱导Ugp1的转录[55]。Msn2/4是具有激活糖代谢和压力响应双重作用的转录因子,在营养物质限制状况下,乙酰-CoA能够刺激细胞增长,Msn2/4可直接结合或刺激激活编码糖酵解酶基因,当Msn2/4缺失时,普遍抑制糖酵解基因表达、乙酰-CoA积累并使细胞进入休眠期[57]。
4 结论和展望TOR信号通路是将环境营养信号和代谢过程关联并维持细胞内稳态的中心调控器。当前,TOR信号通路研究主要集中在哺乳动物细胞和模式菌酿酒酵母中,哺乳动物细胞中主要围绕TOR与疾病发生,以及对肿瘤、癌症等疾病治疗的分子机制。酿酒酵母是最早发现TOR蛋白和研究Rap作用机制的模式生物,哺乳动物TORC1 (mammalian TOR,mTORC1)和酵母TORC1有很多相似调控机制(如激活机制、调控氨基酸合成等[58]),TOR通路中部分重要调控蛋白也具有较高同源性(如Sch9和S6K1等),因此研究真菌酵母细胞TOR信号通路在疾病发生机制探索中可发挥更大的作用。
TORC1感知环境中营养物质和压力信号,由Gtr和Gln两种激活机制激活。根据不同信号调节Sch9激酶和Tap42-Ppase两大支路,由此分别调控下游核糖体合成、自噬、NCR通路、RTG通路以及胁迫响应等细胞生长和代谢的生命过程。相对于TORC1,TORC2的细胞功能研究较少,而TORC2对细胞生长的调控可能是因为TORC2调控细胞骨架肌动蛋白的极化现象、肌动蛋白驱动的内吞作用以及控制质膜稳态[59],其深层次的机制还不清楚。TORC1和TORC2有不同的结构和功能,上下游效应分子一直是研究热点,但两种TOR复合物蛋白之间关联对于细胞生长的调控却鲜有报道。将TORC1和TORC2相关联,共同探究TOR对细胞生长的调控,将为TOR调控细胞生长提供新思路。
此外,TORC1对环境营养信号响应目前仅限于对碳、氮利用(如RTG和NCR通路等),对其下游代谢产物调控非常少。环境营养信号与代谢产物调控是发酵过程主要关注的科学问题,TOR作为真菌细胞生长和代谢的调控中心,通过TOR信号介导调控代谢产物合成,将有助于真菌细胞生长和代谢产物合成机制的理解。
| [1] | Manning BD. Game of TOR-the target of rapamycin rules four kingdoms. The New England Journal of Medicine, 2017, 377(13): 1297-1299. DOI:10.1056/NEJMcibr1709384 |
| [2] | Heitman J, Movva NR, Hiestand PC, Hall MN. FK 506-binding protein proline rotamase is a target for the immunosuppressive agent FK 506 in Saccharomyces cerevisiae. Proceedings of the National Academy of Sciences of the United States of America, 1991, 88(5): 1948-1952. DOI:10.1073/pnas.88.5.1948 |
| [3] | Heitman J, Movva NR, Hall MN. Targets for cell cycle arrest by the immunosuppressant rapamycin in yeast. Science, 1991, 253(5022): 905-909. DOI:10.1126/science.1715094 |
| [4] | Lushchak O, Strilbytska O, Piskovatska V, Storey KB, Koliada A, Vaiserman A. The role of the TOR pathway in mediating the link between nutrition and longevity. Mechanisms of Ageing and Development, 2017, 164: 127-138. DOI:10.1016/j.mad.2017.03.005 |
| [5] | Loewith R, Hall MN. Target of rapamycin (TOR) in nutrient signaling and growth control. Genetics, 2011, 189(4): 1177-1201. DOI:10.1534/genetics.111.133363 |
| [6] | Wullschleger S, Loewith R, Oppliger W, Hall MN. Molecular organization of target of rapamycin complex 2. Journal of Biological Chemistry, 2005, 280(35): 30697-30704. DOI:10.1074/jbc.M505553200 |
| [7] | Fingar DC, Blenis J. Target of rapamycin (TOR):an integrator of nutrient and growth factor signals and coordinator of cell growth and cell cycle progression. Oncogene, 2004, 23(18): 3151-3171. DOI:10.1038/sj.onc.1207542 |
| [8] | Zheng XF, Florentino D, Chen J, Crabtree GR, Schreiber SL. TOR kinase domains are required for two distinct functions, only one of which is inhibited by rapamycin. Cell, 1995, 82(1): 121-130. DOI:10.1016/0092-8674(95)90058-6 |
| [9] | Adami A, García-Álvarez B, Arias-Palomo E, Barford D, Llorca O. Structure of TOR and its complex with KOG1. Molecular Cell, 2007, 27(3): 509-516. DOI:10.1016/j.molcel.2007.05.040 |
| [10] | Loewith R, Jacinto E, Wullschleger S, Lorberg A, Crespo JL, Bonenfant D, Oppliger W, Jenoe P, Hall MN. Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Molecular Cell, 2002, 10(3): 457-468. DOI:10.1016/S1097-2765(02)00636-6 |
| [11] | Reinke A, Anderson S, McCaffery JM, Yates Ⅲ J, Aronova S, Chu S, Fairclough S, Iverson C, Wedaman KP, Powers T. TOR complex 1 includes a novel component, Tco89p (YPL180w), and cooperates with Ssd1p to maintain cellular integrity in Saccharomyces cerevisiae. Journal of Biological Chemistry, 2004, 279(15): 14752-14762. DOI:10.1074/jbc.M313062200 |
| [12] | Wedaman KP, Reinke A, Anderson S, Yates Ⅲ J, McCaffery JM, Powers T. Tor kinases are in distinct membrane-associated protein complexes in Saccharomyces cerevisiae. Molecular Biology of the Cell, 2003, 14(3): 1204-1220. DOI:10.1091/mbc.e02-09-0609 |
| [13] | Kira S, Kumano Y, Ukai H, Takeda E, Matsuura A, Noda T. Dynamic relocation of the TORC1-Gtr1/2-Ego1/2/3 complex is regulated by Gtr1 and Gtr2. Molecular Biology of the Cell, 2016, 27(2): 382-396. DOI:10.1091/mbc.e15-07-0470 |
| [14] | Tanigawa M, Maeda T. An in vitro TORC1 kinase assay that recapitulates the Gtr-independent glutamine-responsive TORC1 activation mechanism on yeast vacuoles. Molecular and Cellular Biology, 2017, 37(14). DOI:10.1128/MCB.00075-17 |
| [15] | Hallett JEH, Luo XX, Capaldi AP. State transitions in the TORC1 signaling pathway and information processing in Saccharomyces cerevisiae. Genetics, 2014, 198(2): 773-786. DOI:10.1534/genetics.114.168369 |
| [16] | Wei Y, Zheng XFS. Nutritional control of cell growth via TOR signaling in budding yeast. Methods in Molecular Biology, 2011, 759: 307-319. DOI:10.1007/978-1-61779-173-4 |
| [17] | Urban J, Soulard A, Huber A, Lippman S, Mukhopadhyay D, Deloche O, Wanke V, Anrather D, Ammerer G, Riezman H, Broach JR, De Virgilio C, Hall MN, Loewith R. Sch9 is a major target of TORC1 in Saccharomyces cerevisiae. Molecular Cell, 2007, 26(5): 663-674. DOI:10.1016/j.molcel.2007.04.020 |
| [18] | González A, Shimobayashi M, Eisenberg T, Merle DA, Pendl T, Hall MN, Moustafa T. TORC1 promotes phosphorylation of ribosomal protein S6 via the AGC kinase Ypk3 in Saccharomyces cerevisiae. PLoS One, 2015, 10(3): e0120250. DOI:10.1371/journal.pone.0120250 |
| [19] | Gu Q, Zhang CQ, Yu FW, Yin YN, Shim WB, Ma ZH. Protein kinase FgSch9 serves as a mediator of the target of rapamycin and high osmolarity glycerol pathways and regulates multiple stress responses and secondary metabolism in Fusarium graminearum. Environmental Microbiology, 2015, 17(8): 2661-2676. DOI:10.1111/1462-2920.12522 |
| [20] | Alves de Castro P, Dos Reis TF, Dolan SK, Oliveira Manfiolli A, Brown NA, Jones GW, Doyle S, Riaño-Pachón DM, Squina FM, Caldana C, Singh A, del Poeta M, Hagiwara D, Silva-Rocha R, Goldman GH. The Aspergillus fumigatus SchASCH9kinase modulates SakAHOG1MAP kinase activity and it is essential for virulence. Molecular Microbiology, 2016, 102(4): 642-671. DOI:10.1111/mmi.2016.102.issue-4 |
| [21] | Huber A, French SL, Tekotte H, Yerlikaya S, Stahl M, Perepelkina MP, Tyers M, Rougemont J, Beyer AL, Loewith R. Sch9 regulates ribosome biogenesis via Stb3, Dot6 and Tod6 and the histone deacetylase complex RPD3L. The EMBO Journal, 2011, 30(15): 3052-3064. DOI:10.1038/emboj.2011.221 |
| [22] | Broach JR. Nutritional control of growth and development in yeast. Genetics, 2012, 192(1): 73-105. DOI:10.1534/genetics.111.135731 |
| [23] | Jorgensen P, Rupeš I, Sharom JR, Schneper L, Broach JR, Tyers M. A dynamic transcriptional network communicates growth potential to ribosome synthesis and critical cell size. Genes & Development, 2004, 18(20): 2491-2505. |
| [24] | Cai Y, Wei YH. Distinct regulation of Maf1 for lifespan extension by Protein kinase A and Sch9. Aging, 2015, 7(2): 133-143. DOI:10.18632/aging.v7i2 |
| [25] | Wei YH, Tsang CK, Zheng XFS. Mechanisms of regulation of RNA polymerase Ⅲ-dependent transcription by TORC1. The EMBO Journal, 2009, 28(15): 2220-2230. DOI:10.1038/emboj.2009.179 |
| [26] | Chymkowitch P, Nguéa PA, Aanes H, Robertson J, Klungland A, Enserink JM. TORC1-dependent sumoylation of Rpc82 promotes RNA polymerase Ⅲ assembly and activity. Proceedings of the National Academy of Sciences of the United States of America, 2017, 114(5): 1039-1044. DOI:10.1073/pnas.1615093114 |
| [27] | Li H, Tsang CK, Watkins M, Bertram PG, Zheng XFS. Nutrient regulates Tor1 nuclear localization and association with rDNA promoter. Nature, 2006, 442(7106): 1058-1061. DOI:10.1038/nature05020 |
| [28] | Torreira E, Louro JA, Pazos I, González-Polo N, Gil-Carton D, Duran AG, Tosi S, Gallego O, Calvo O, Fernández-Tornero C. The dynamic assembly of distinct RNA polymerase I complexes modulates rDNA transcription. eLife, 2017, 6: e20832. DOI:10.7554/eLife.20832 |
| [29] | Noda T. Regulation of Autophagy through TORC1 and mTORC1. Biomolecules, 2017, 7(3): 52. |
| [30] | Kamada Y, Yoshino K, Kondo C, Kawamata T, Oshiro N, Yonezawa K, Ohsumi Y. Tor directly controls the Atg1 kinase complex to regulate autophagy. Molecular and Cellular Biology, 2010, 30(4): 1049-1058. DOI:10.1128/MCB.01344-09 |
| [31] | Shin CS, Huh WK. Bidirectional regulation between TORC1 and autophagy in Saccharomyces cerevisiae. Autophagy, 2011, 7(8): 854-862. DOI:10.4161/auto.7.8.15696 |
| [32] | Yeasmin AM, Waliullah TM, Kondo A, Kaneko A, Koike N, Ushimaru T. Orchestrated action of PP2A antagonizes Atg13 phosphorylation and promotes autophagy after the inactivation of TORC1. PLoS One, 2016, 11(12): e0166636. DOI:10.1371/journal.pone.0166636 |
| [33] | Swer PB, Lohia R, Saran S. Analysis of rapamycin induced autophagy in Dictyostelium discoideum. Indian Journal of Experimental Biology, 2014, 52(4): 295-304. |
| [34] | Ding BB, Zhong Q. Zinc deficiency:An unexpected trigger for autophagy. The Journal of Biological Chemistry, 2017, 292(20): 8531-8532. DOI:10.1074/jbc.H116.762948 |
| [35] | Umekawa M, Ujihara M, Nakai D, Takematsu H, Wakayama M. Ecm33 is a novel factor involved in efficient glucose uptake for nutrition-responsive TORC1 signaling in yeast. FEBS Letters, 2017, 591(22): 3721-3729. DOI:10.1002/feb2.2017.591.issue-22 |
| [36] | Conrad M, Schothorst J, Kankipati HN, Van Zeebroeck G, Rubio-Texeira M, Thevelein JM. Nutrient sensing and signaling in the yeast Saccharomyces cerevisiae. FEMS Microbiology Reviews, 2014, 38(2): 254-299. DOI:10.1111/1574-6976.12065 |
| [37] | Pfannmüller A, Leufken J, Studt L, Michielse CB, Sieber CMK, Güldener U, Hawat S, Hippler M, Fufezan C, Tudzynski B. Comparative transcriptome and proteome analysis reveals a global impact of the nitrogen regulators AreA and AreB on secondary metabolism in Fusarium fujikuroi. PLoS One, 2017, 12(4): e0176194. DOI:10.1371/journal.pone.0176194 |
| [38] | Georis I, Tate JJ, Cooper TG, Dubois E. Nitrogen-responsive regulation of GATA protein family activators Gln3 and Gat1 occurs by two distinct pathways, one inhibited by rapamycin and the other by methionine sulfoximine. Journal of Biological Chemistry, 2011, 286(52): 44897-44912. DOI:10.1074/jbc.M111.290577 |
| [39] | Georis I, Tate JJ, Cooper TG, Dubois E. Tor pathway control of the nitrogen-responsive DAL5 gene bifurcates at the level of Gln3 and Gat1 regulation in Saccharomyces cerevisiae. Journal of Biological Chemistry, 2008, 283(14): 8919-8929. DOI:10.1074/jbc.M708811200 |
| [40] | Chen XR, Wang ZY, Guo XN, Liu S, He XP. Regulation of general amino acid permeases Gap1p, GATA transcription factors Gln3p and Gat1p on 2-phenylethanol biosynthesis via Ehrlich pathway. Journal of Biotechnology, 2017, 242: 83-91. DOI:10.1016/j.jbiotec.2016.11.028 |
| [41] | Numamoto M, Tagami S, Ueda Y, Imabeppu Y, Sasano Y, Sugiyama M, Maekawa H, Harashima S. Nuclear localization domains of GATA activator Gln3 are required for transcription of target genes through dephosphorylation in Saccharomyces cerevisiae. Journal of Bioscience and Bioengineering, 2015, 120(2): 121-127. DOI:10.1016/j.jbiosc.2014.12.017 |
| [42] | Pérez-Delos Santos FJ, Riego-Ruiz L. Gln3 is a main regulator of nitrogen assimilation in Candida glabrata. Microbiology, 2016, 162(8): 1490-1499. DOI:10.1099/mic.0.000312 |
| [43] | Baldin C, Valiante V, Krüger T, Schafferer L, Haas H, Kniemeyer O, Brakhage AA. Comparative proteomics of a tor inducible Aspergillus fumigatus mutant reveals involvement of the Tor kinase in iron regulation. Proteomics, 2015, 15(13): 2230-2243. DOI:10.1002/pmic.v15.13 |
| [44] | Wang YK, Song XD, Zhang YJ, Wang BC, Zou X. Effects of nitrogen availability on polymalic acid biosynthesis in the yeast-like fungus Aureobasidium pullulans. Microbial Cell Factories, 2016, 15(1): 146. DOI:10.1186/s12934-016-0547-y |
| [45] | Liu NN, Flanagan PR, Zeng JM, Jani NM, Cardenas ME, Moran GP, Köhler JR. Phosphate is the third nutrient monitored by TOR in Candida albicans and provides a target for fungal-specific indirect TOR inhibition. Proceedings of the National Academy of Sciences of the United States of America, 2017, 114(24): 6346-6351. DOI:10.1073/pnas.1617799114 |
| [46] | da Cunha FM, Torelli NQ, Kowaltowski AJ. Mitochondrial retrograde signaling:triggers, pathways, and outcomes. Oxidative Medicine and Cellular Longevity, 2015, 2015: 482582. |
| [47] | Liu ZC, Butow RA. Mitochondrial retrograde signaling. Annual Review of Genetics, 2006, 40: 159-185. DOI:10.1146/annurev.genet.40.110405.090613 |
| [48] | Peters TW, Miller AW, Tourette C, Agren H, Hubbard A, Hughes RE. Genomic analysis of ATP efflux in Saccharomyces cerevisiae. G3-Genes Genomes Genetics, 2016, 6(1): 161-170. |
| [49] | Rios-Anjos RM, de Lima Camandona V, Bleicher L, Ferreira-Junior JR. Structural and functional mapping of Rtg2p determinants involved in retrograde signaling and aging of Saccharomyces cerevisiae. PLoS One, 2017, 12(5): e0177090. DOI:10.1371/journal.pone.0177090 |
| [50] | Eisenberg-Bord M, Schuldiner M. Ground control to major TOM:mitochondria-nucleus communication. FEBS Journal, 2017, 284(2): 196-210. DOI:10.1111/febs.13778 |
| [51] | González B, Mas A, Beltran G, Cullen PJ, Torija MJ. Role of mitochondrial retrograde pathway in regulating ethanol-inducible filamentous growth in yeast. Frontiers in Physiology, 2017, 8: 148. |
| [52] | Madeira JB, Masuda CA, Maya-Monteiro CM, Matos GS, Montero-Lomeli M, Bozaquel-Morais BL. TORC1 inhibition induces lipid droplet replenishment in yeast. Molecular and Cellular Biology, 2015, 35(4): 737-746. DOI:10.1128/MCB.01314-14 |
| [53] | Torelli NQ, Ferreira-Júnior JR, Kowaltowski AJ, da Cunha FM. RTG1-and RTG2-dependent retrograde signaling controls mitochondrial activity and stress resistance in Saccharomyces cerevisiae. Free Radical Biology and Medicine, 2015, 81: 30-37. DOI:10.1016/j.freeradbiomed.2014.12.025 |
| [54] | Laera L, Guaragnella N, Ždralević M, Marzulli D, Liu ZC, Giannattasio S. The transcription factors ADR1 or CAT8 are required for RTG pathway activation and evasion from yeast acetic acid-induced programmed cell death in raffinose. Microbial Cell, 2016, 3(12): 621-631. DOI:10.15698/mic |
| [55] | Yi DG, Huh WK. PKA, PHO and stress response pathways regulate the expression of UDP-glucose pyrophosphorylase through Msn2/4 in budding yeast. FEBS Letters, 2015, 589(18): 2409-2416. DOI:10.1016/j.febslet.2015.07.015 |
| [56] | Morano KA, Grant CM, Moye-Rowley WS. The response to heat shock and oxidative stress in Saccharomyces cerevisiae. Genetics, 2012, 190(4): 1157-1195. DOI:10.1534/genetics.111.128033 |
| [57] | Kuang Z, Pinglay S, Ji HK, Boeke JD. Msn2/4 regulate expression of glycolytic enzymes and control transition from quiescence to growth. eLife, 2017, 6: e29938. DOI:10.7554/eLife.29938 |
| [58] | González A, Hall MN. Nutrient sensing and TOR signaling in yeast and mammals. The EMBO Journal, 2017, 36(4): 397-408. DOI:10.15252/embj.201696010 |
| [59] | Roelants FM, Leskoske KL, Martinez Marshall MN, Locke MN, Thorner J. The TORC2-dependent signaling network in the yeast Saccharomyces cerevisiae. Biomolecules, 2017, 7(3): 66. |
 2018, Vol. 58
2018, Vol. 58