中国科学院微生物研究所,中国微生物学会,中国菌物学会
文章信息
- 徐冬怡, 蒋佳利, 方仁东. 2020
- Dongyi Xu, Jiali Jiang, Rendong Fang. 2020
- 肺炎链球菌与宿主炎症小体的相互作用机制
- Mechanisms underlying the interaction between Streptococcus pneumoniae and host inflammasomes
- 微生物学报, 60(10): 2111-2121
- Acta Microbiologica Sinica, 60(10): 2111-2121
-
文章历史
- 收稿日期:2019-11-17
- 修回日期:2020-01-07
- 网络出版日期:2020-02-28
2. 西南大学医学研究院免疫学研究中心, 重庆 402460
2. Immunology Research Center, Medical Research Institute, Southwest University, Chongqing 402460, China
肺炎链球菌(Streptococcus pneumoniae)是一种条件性致病的革兰阳性胞外菌,经常定植于人上呼吸道器官的黏膜中。这种无症状定植在宿主免疫状态低下和强毒血清型感染时,可能发展为侵袭性疾病,如社区获得性肺炎、败血症、脑膜炎、中耳炎等。肺炎链球菌所引起的疾病有较高的死亡率,在儿童患者中尤为突出,每年死亡人数约计120万[1]。迄今为止,根据关键毒力因子荚膜多糖结构划分的肺炎链球菌血清型约有98种,其中一些血清型的肺炎链球菌可以在鼻、咽部定植,所引起的疾病具有较高的死亡率。其他血清型常常在侵袭性疾病中发现,但其引起的疾病死亡率较低[2-3]。肺炎链球菌溶血素(pneumolysin,PLY)是胆固醇依赖性细胞毒素家族成员之一。大多数肺炎链球菌的PLY通过与胞膜上的胆固醇结合使细胞膜形成穿孔进而导致细胞溶解,具有细胞毒性[3]。PLY在引发疾病中的关键性作用已在各种肺炎链球菌疾病模型中得到证实,它是肺炎链球菌引起机体先天免疫反应的成分之一[4]。
模式识别受体(pattern recognition receptors,PRRs)可识别病原体相关分子模式(pathogen- associated molecular patterns,PAMPs)的保守微生物组分,这是机体先天免疫系统的第一道屏障[5]。目前已经发现的PRRs家族有5类,包括C型凝集素受体(C-type lectin receptors,CLRs)、RIG-I样受体(RIG-I-like receptors,RLRs)、Toll样受体(Toll-like receptors,TLRs)、NOD样受体(NOD-like receptors,NLRs)和AIM2样受体(AIM2-like receptors,ALRs)家族,不同的PRRs分布在机体的不同部位,识别不同的PAMPs,最终诱导机体产生免疫应答[6]。炎症小体是一种多蛋白复合物,由受体蛋白、衔接蛋白和效应蛋白组成[7]。炎症小体的受体蛋白大都属于NLRs家族和ALRs家族,根据受体蛋白组成的不同,目前研究较多的是NLRP3、NLRP1、NLRC4、NLRP6和AIM2炎症小体,NLRP2、NLRP7、NLRP12等炎症小体也有报道[8]。PRRs识别PAMPs后借助衔接蛋白凋亡相关斑点样蛋白(apoptosis-associated speck-like protein containing a CARD,ASC)来募集和激活半胱天冬酶1前体(pro-caspase-1)和/或半胱天冬酶11前体(pro-caspase-11)效应蛋白,并使caspase-1和/或caspase-11的前体切割为成熟形式并发挥作用[9],进而调控细胞程序性死亡即细胞焦亡(pyroptosis)以及细胞因子IL-1β和IL-18的成熟与分泌[10]。最新研究表明,gasdermin D(GSDMD)也是半胱天冬酶1的靶标之一,GSDMD裂解后作用于胞膜形成gasdermin孔,最终导致膜的破裂并参与到细胞焦亡的过程中[11]。
越来越多的研究表明,肺炎链球菌入侵宿主后可引起不同炎症小体的活化,而不同炎症小体组分的缺失也会引起机体病程的改变。本文将结合本团队的相关研究工作,对肺炎链球菌感染过程中炎症小体的激活、炎症小体在宿主抗肺炎链球菌中的作用、肺炎链球菌对宿主炎症小体的逃避策略等相关研究进展进行综合阐述。
1 肺炎链球菌感染过程中炎症小体的激活Tschopp等首次提出,典型的炎症小体是细胞溶质蛋白复合物,它是募集和活化pro-caspase-1的平台[12]。而参与肺炎链球菌识别的PRRs主要为NLRs家族的NLRP3炎症小体和ALRs家族的AIM2炎症小体[6],不同的炎症小体所识别的肺炎链球菌的配体各不相同。
1.1 肺炎链球菌激活炎症小体炎症小体的激活过程包括炎症小体识别肺炎链球菌以及NF-κB介导的IL-1β前体的表达上调[13]。NLRP3炎症小体对多种微生物和内源分子诱导的细胞紊乱均有反应[14-15],很多研究证实肺炎链球菌的PLY可以引起NLRP3炎症小体的激活[16]。Shoma等已证明,重组的PLY蛋白可以诱导巨噬细胞中IL-18、IL-1β以及IL-1α的分泌[17]。本团队最近研究证实,在肺炎链球菌感染中性粒细胞过程中,PLY参与NLRP3炎症小体的活化[18]。然而,肺炎链球菌PLY激活NLRP3炎症小体的具体机制尚未研究清楚,推测其可能为间接识别机制。在肺炎链球菌感染细胞的过程中,PLY的成孔作用可导致溶酶体破裂和细菌RNA及其他内源性分子释放到胞质,因此PLY活化NLRP3炎症小体的确切机制可能涉及到钾离子外流、溶酶体破裂或RNA的激活等。Gupta等证明,化脓性链球菌的RNA为小鼠巨噬细胞中NLRP3炎症小体的直接激活剂[19],肺炎链球菌的RNA是否亦可激活NLRP3有待进一步研究。
多项研究表明,在肺炎链球菌感染细胞的过程中,AIM2能识别并结合肺炎链球菌的dsDNA,进而激活AIM2炎症小体的组装[20-21]。AIM2识别释放到细胞质中的微生物DNA,由于肺炎链球菌是胞外寄生菌,在宿主免疫过程中被巨噬细胞吞噬后会迅速死亡[22],因此,AIM2识别肺炎链球菌还依赖于PLY的胞膜成孔作用和LytA的溶菌作用[22]。此外,TLR-9也能识别肺炎链球菌的DNA,并通过细胞溶质感应器检测,最后通过衔接分子STING发出信号[23]。这说明AIM2炎症小体可能还与其他DNA受体蛋白相关的信号通路存在交叉作用,他们之间的相互作用机制还有待进一步的探索。
已有研究表明,肺炎链球菌感染任何模式细胞均不会引起NLRC4炎症小体的激活[24]。但在NLRP3和AIM2炎症小体之外,是否还有其他炎症小体参与肺炎链球菌的识别还有待进一步研究。Hara等[25]研究表明在一些革兰阳性菌感染巨噬细胞时,NLRP6介导的caspase-11作用于caspase-1的上游切割IL-1β和IL-18的前体。NLRP6炎症小体可通过识别一些革兰阳性菌的磷壁酸(lipoteichoic acid,LTA)形成复合物,该复合物能够引起ASC的寡聚化进而诱导下游细胞因子的成熟与分泌。而肺炎链球菌感染是否诱导NLRP6炎症小体的激活有待进一步研究。
肺炎链球菌感染诱导炎症小体的激活可引起caspase-1的活化和下游细胞因子的分泌。本团队已证明,在NLRP3–/–、AIM2–/–或ASC–/–小鼠的腹腔巨噬细胞中,NLRP3、AIM2、ASC缺失时会导致caspase-1的成熟形式减少及IL-1β、IL-18的分泌水平显著降低[20],当caspase-1缺失时同样会引起IL-1β和IL-18分泌量的下降[26]。在小鼠中性粒细胞[18]及人类中性粒细胞[27]中,肺炎链球菌可以激活NLRP3炎症小体的组装,使caspase-1活化并参与IL-1β的成熟与分泌,但AIM2炎症小体不参与该过程。在小胶质细胞、树突状细胞、人类单核细胞及小鼠单核细胞模型中均发现,肺炎链球菌感染后caspase-1的活化呈NLRP3依赖性,且诱导IL-1β和IL-18的成熟与分泌[28-29] (图 1)。
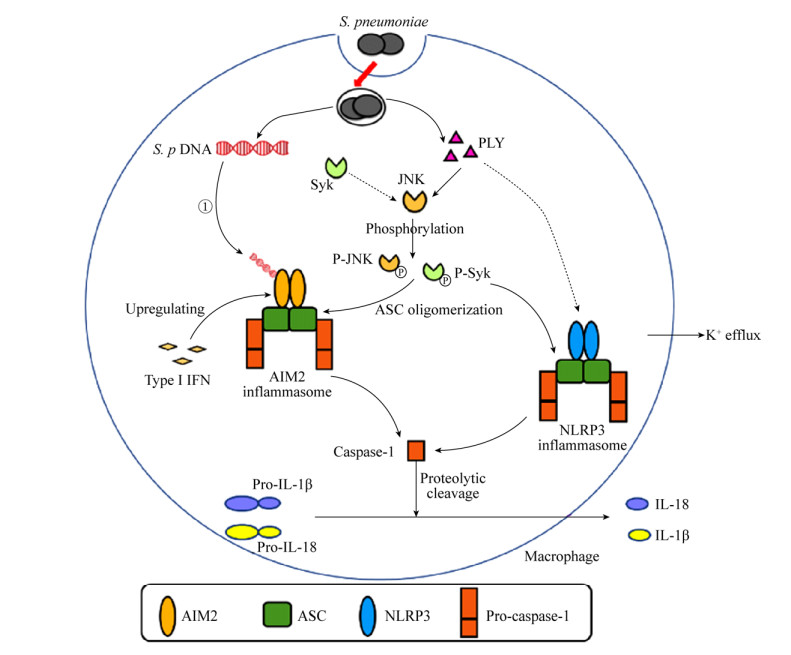
|
| 图 1 肺炎链球菌感染诱导炎症小体激活的机制 Figure 1 Mechanism underlying inflammasome activation induced by S. pneumoniae infection. ① Streptococcus pneumoniae DNA spills from phagosomes into cytoplasm and activates AIM2 inflammasome; ② Pneumolysin and K+ efflux activate NLRP3 inflammasome; ③ Activation of AIM2 and NLRP3 inflammasomes cause cleavage of caspase-1 from precursor to its mature form; ④ Pro-IL-1β and pro-IL-18 are cleaved by actived caspase-1 into bioactive form and secreted. |
1.2 肺炎链球菌诱导炎症小体激活的调控机制
在肺炎链球菌感染机体过程中,炎症小体的活化受多方面的调控。一方面,一些物质可以引起炎症小体的正调控。肺炎链球菌在感染巨噬细胞和中性粒细胞时,会导致细胞内钾离子浓度降低即钾离子外流,介导ASC寡聚化[30]。Syk和JNK对炎症小体的激活也发挥着重要作用。本团队研究发现,用JNK抑制剂(SP600125)及Syk抑制剂(R406)预处理的巨噬细胞在感染肺炎链球菌后IL-1β的分泌量、ASC寡聚化及caspase-1活化形式均显著降低,证明JNK、Syk通过介导ASC寡聚化参与调控肺炎链球菌感染的巨噬细胞所诱导的炎症小体的活化[31]。本团队在中性粒细胞中发现,仅有JNK参与肺炎链球菌感染后炎症小体的活化,Syk没有参与,且中性粒细胞中富含丝氨酸蛋白酶类,该酶类同样可以调节ASC分子寡聚化[18]。NLRP3炎症小体的启动需要脂肪酸的合成,Moon等证明,在NLRP3炎症小体激活过程中,NADPH氧化酶4(NOX4)依赖的脂肪酸氧化是NLRP3依赖的ASC寡聚化所必需的[32]。干扰素(interferons,IFNs)作为一类具有免疫调节及抗肿瘤功能的因子,在肺炎链球菌感染宿主时也发挥着不可或缺的作用[33]。早在1986年就发现,IFN可调控肺炎链球菌对宿主的感染[34]。本团队前期利用IFNAR1–/–动物模型证明,Ⅰ型IFN的信号传导对于肺炎链球菌感染所引起的AIM2的上调及IL-18的分泌是必需的,但这一过程不影响巨噬细胞摄取和杀死肺炎链球菌,且Ⅰ型IFN不会引起NLRP3表达的上调[35] (图 1)。
另一方面,相关文献已经报道了几种可以负性调节炎症小体的物质,例如几丁质酶3样蛋白1 (Chi3l1)是由肺炎链球菌诱导所产生的一种保守的几丁质样蛋白原型。有研究表明,Chi3l1通过抑制caspase-1依赖型巨噬细胞焦亡来增强巨噬细胞的杀伤能力,与野生型对照组相比,肺炎链球菌感染Chi3l1缺失小鼠会表现出过度的肺损伤、炎症和出血,但NLRP3、IL-1β及IL-18分泌量均升高,表明Chi3l1可以通过抑制NLRP3炎症小体的活化,避免炎性细胞因子的过表达,达到保护机体的作用[36]。在肺炎链球菌自身分泌的负性调节炎症小体的因子中,过氧化氢(H2O2)可抑制炎症小体,进而削弱免疫系统并导致肺炎[37],而活性氧(ROS)在炎症小体中的作用目前还有较大争议[38]。
2 炎症小体在抗肺炎链球菌中的作用当肺炎链球菌感染宿主时,会发生肺炎、脑膜炎等症状,很多学者也探究了炎症小体在肺炎链球菌感染不同动物模型中所发挥的不同作用。首先本团队早期发现,NLRP3–/–、ASC–/–动物显示出对肺炎链球菌的高度敏感性,且肺炎链球菌在肺部的细菌定植量明显增高,肺部出现更加明显的病理现象,死亡率也显著升高,同时一些细胞因子如IL-1β、IL-18的分泌水平会降低[21]。随后本团队又发现,AIM2缺陷型动物也具有与NLRP3–/–、ASC–/–动物相似的结果[39]。但在肺炎链球菌脑膜炎中,NLRP3炎症小体不具有宿主保护作用。感染肺炎链球菌的ASC–/–动物的颅内压显著低于野生型动物,且NLRP3–/–、ASC–/–动物的病理及临床评分都优于野生型动物[40]。在该动物模型中发现,炎症小体对脑膜炎病程的推进作用是由于IL-18的产生可诱导IFN-γ的分泌,是Ⅱ型IFN在影响病情的发展[41]。在脑脊液中的炎症相关细胞因子与肺炎链球菌脑膜炎的并发症及不良预后有关,相关炎症小体组分的缺失也会使炎症现象大幅下降[42-43]。有研究发现,IL-1β、IL-18缺陷型小鼠比野生型小鼠更易患肺炎链球菌肺炎[44]。此外IL-1的产生对于肺炎链球菌在上呼吸道定植模型中的细菌限制和传播至关重要,但IL-1并没有参与机体的适应性免疫应答,且炎症小体-IL-1β途径在肺炎链球菌感染过程中的保护作用可能部分取决于中性粒细胞和/或巨噬细胞的募集[45]。
在肺泡上皮细胞中,肺炎链球菌感染的NLRP3–/–动物中受损的肺屏障功能与测试时间点的IL-1β和IL-18分泌水平降低无关,且caspase-1、IL-1β和IL-18缺失的小鼠单层肺泡上皮细胞在PLY处理后未显示出屏障功能障碍加重的现象,表明NLRP3在保护肺炎链球菌感染的肺上皮细胞屏障功能不依赖ASC、caspase-1、IL-1β和IL-18[46]。本团队最新研究表明,caspase-1/11–/–动物的生存曲线、肺炎链球菌细菌定植量及肺部病理变化与野生型动物相比没有明显差异,且ASC和NLRP3可通过涉及STAT6-SPDEF途径的非依赖炎症小体的机制促进气道黏膜的先天免疫,说明NLRP3可以通过多种途径保护宿主免受肺炎链球菌的侵害。在感染过程中,ASC和NLRP3维持转录因子SPDEF的表达,这可以促进黏膜防御基因如Tff2、Reg3γ、Bpifal的表达。而STAT6作为SPDEF表达的关键调节因子,它的激活也依赖于ASC和NLRP3[47] (图 2)。这个发现揭示了NLRP3的caspase-1非依赖性作用新机制,将有助于控制微生物感染,同时为气道黏膜中先天免疫稳态的深入研究奠定基础。综上所述,NLRP3可通过多种机制保护宿主不受肺炎链球菌的侵害。
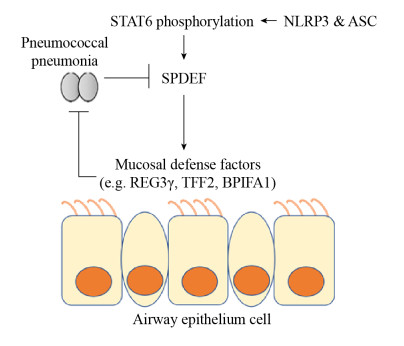
|
| 图 2 NLRP3通过caspase-1非依赖途径保护宿主抗肺炎链球菌感染的机制 Figure 2 NLRP3 protects the host against S. pneumoniae infection through a caspase-1-independent mechanism. NLRP3 and ASC maintain the production of the transcription factor SPDEF by inducing the phosphorylation of STAT6, and promoting the expression of mucosal defense genes such as Tff2, Reg3γ and Bpifal, and protecting the host against S. pneumoniae. The whole process is independent of the role of caspase-1. |
总之,以上研究表明炎症小体一般情况下可以协助机体的其他免疫反应抵御肺炎链球菌的感染,在先天免疫中发挥着重要的作用。但是在不同的活化部位可能也会对宿主产生不利的后果,有助于加剧炎症反应,引起组织更严重的病理变化。
3 肺炎链球菌逃避宿主炎症小体识别的机制在漫长的选择进化中,宿主炎症小体的作用会对病原体产生选择压力,使病原体产生抑制炎症小体作用的自我保护机制[7]。肺炎链球菌的关键性毒力因子PLY对宿主细胞具有细胞毒性,可抑制呼吸道上皮细胞的纤毛痉挛,激活经典的补体系统,并刺激炎症小体从而影响机体的免疫系统,同时还可参与生物被膜的形成和引发组织损伤[4],在增强肺炎链球菌侵袭力的同时引起宿主损伤[3]。研究表明,在人类树突状细胞中PLY的表达可帮助细菌逃避模式识别,抑制树突细胞中细胞因子的产生[48]。
PLY的氨基酸序列最初被认为在肺炎链球菌的所有血清型中都是相对保守的,随着时间的推移,其序列变化不大[49]。但是由于选择压力,肺炎链球菌能产生突变。临床上一些重要的肺炎链球菌菌株,如新发现的血清1型MLST306菌株,由于其与肺炎胸腔积液和脓胸有关,再加上它们与小型或封闭社区的疾病暴发有关,因而成为最常见的侵袭性肺炎链球菌之一[50]。Kirkham等[51]在1型ST306分离株中首次发现了等位基因5。有趣的是,ST306表达非细胞溶解性PLY突变体,该分离株中非溶血性PLY仍可显著增加肺炎链球菌的毒力[52],并保持其与含胆固醇膜及寡聚体结合的能力,但它不具有成孔作用[51]。Fatykhova等[53]发现,表达肺炎链球菌PLY等位基因5的菌株确实可以逃避炎症小体的识别,如血清型1 ST306、ST228、ST617等和血清型8的ST53、ST578、ST835、ST1110和ST1722等[52]。用ST306或ST53感染人类单核细胞和小鼠骨髓来源巨噬细胞,发现与其他肺炎链球菌感染所引起的强烈反应相比,PLY等位基因5只能诱导少量IL-1β的产生[29]。非溶血性PLY的表达可能使血清型1细菌对宿主的损害较小,有助于解释为什么血清1型肺炎链球菌感染与其他肺炎链球菌感染所引起的疾病相比具有较低的病死率[54],并且与侵入性肺炎球菌疾病的暴发有关[55]。由此可以推测,具有PLY等位基因5的菌株对肺炎链球菌导致的炎症小体依赖的免疫逃逸机制可能有助于它们进入宿主无菌部位并增强对宿主侵袭性的感染能力,例如MLST306与其他肺炎链球菌相比,其在鼻咽部定植的时间可能会延长[56]。值得注意的是,溶血性PLY的表达还是STING依赖途径通过NOD2和DNA检测肺炎链球菌肽聚糖所必需的[57]。因此,逃避这些途径的识别也可能增加血清型1肺炎球菌的相对特异性。总之,不同血清型的肺炎链球菌激活NLRP3炎症小体的能力有差异,并且重要的血清1型和8型菌株可通过这种先天免疫传感机制逃避识别[53]。
4 小结宿主炎症小体是机体免疫系统感知潜在病原微生物的关键参与者,近年来成为研究热点。综上所述,大多数肺炎链球菌感染机体能引起NLRP3、AIM2炎症小体的激活[58],但是肺炎链球菌直接激活NLRP3炎症小体的作用分子目前尚不清楚,推测可能为肺炎链球菌RNA,该过程是否还有其他炎症小体的参与还有待进一步研究。在长期进化过程中,一些肺炎链球菌菌株可通过PLY非溶血性突变体逃避炎症小体的免疫检测[59]。以后可能还会发现更多肺炎链球菌对炎症小体的逃避机制。了解这些机制对疾病的预防和控制有着重要意义。在体内感染肺炎链球菌的情况下,NLRP3和ASC通过caspase-1非依赖的途径诱导宿主防御因子抗感染的现象是一个重要发现,但是这些宿主防御因子Tff2、Reg3γ、Bpifal等是如何抗肺炎链球菌感染的?是否还受其他因素的调控?其详细机制还有待进一步研究。未来,这些基础研究工作的突破将有望对肺炎链球菌感染引起的相关疾病的预防和治疗,包括疫苗研制、药物靶标选择等提供新的思路。
| [1] | van der Poll T, Opal SM. Pathogenesis, treatment, and prevention of pneumococcal pneumonia. The Lancet, 2009, 374(9700): 1543-1556. DOI:10.1016/S0140-6736(09)61114-4 |
| [2] | Koppe U, Suttorp N, Opitz B. Recognition of Streptococcus pneumoniae by the innate immune system. Cellular Microbiology, 2012, 14(4): 460-466. DOI:10.1111/j.1462-5822.2011.01746.x |
| [3] | Mitchell AM, Mitchell TJ. Streptococcus pneumoniae:virulence factors and variation. Clinical Microbiology and Infection, 2010, 16(5): 411-418. DOI:10.1111/j.1469-0691.2010.03183.x |
| [4] | Shak JR, Ludewick HP, Howery KE, Sakai F, Yi H, Harvey RM, Paton JC, Klugman KP, Vidal JE. Novel role for the Streptococcus pneumoniae toxin pneumolysin in the assembly of biofilms. mBio, 2013, 4(5): 00655-13. |
| [5] | Lamkanfi M, Dixit VM. Mechanisms and functions of inflammasomes. Cell, 2014, 157(5): 1013-1022. DOI:10.1016/j.cell.2014.04.007 |
| [6] | Man SM, Kanneganti TD. Regulation of inflammasome activation. Immunological Reviews, 2015, 265(1): 6-21. |
| [7] | Brewer SM, Brubaker SW, Monack DM. Host inflammasome defense mechanisms and bacterial pathogen evasion strategies. Current Opinion in Immunology, 2019, 60: 63-70. DOI:10.1016/j.coi.2019.05.001 |
| [8] | Lupfer C, Kanneganti TD. Unsolved mysteries in NLR biology. Frontiers in Immunology, 2013, 4: 285. |
| [9] | Broz P. Recognition of intracellular bacteria by inflammasomes. Microbiology Spectrum, 2019, 7(2). DOI:10.1128/microbiolspec.BAI-0003-2019 |
| [10] | Kayagaki N, Warming S, Lamkanfi M, Vande Walle L, Louie S, Dong J, Newton K, Qu Y, Liu JF, Heldens S, Zhang J, Lee WP, Roose-Girma M, Dixit VM. Non-canonical inflammasome activation targets caspase-11. Nature, 2011, 479(7371): 117-121. DOI:10.1038/nature10558 |
| [11] | Shin S, Brodsky IE. The inflammasome:Learning from bacterial evasion strategies. Seminars in Immunology, 2015, 27(2): 102-110. DOI:10.1016/j.smim.2015.03.006 |
| [12] | Martinon F, Burns K, Tschopp J. The inflammasome:A molecular platform triggering activation of inflammatory caspases and processing of proIL-β. Molecular Cell, 2002, 10(2): 417-426. DOI:10.1016/S1097-2765(02)00599-3 |
| [13] | Bauernfeind FG, Horvath G, Stutz A, Alnemri ES, MacDonald K, Speert D, Fernandes-Alnemri T, Wu JH, Monks BG, Fitzgerald KA, Hornung V, Latz E. Cutting edge:NF-κB activating pattern recognition and cytokine receptors license NLRP3 inflammasome activation by regulating NLRP3 expression. Journal of Immunology, 2009, 183(2): 787-791. DOI:10.4049/jimmunol.0901363 |
| [14] | Halle A, Hornung V, Petzold GC, Stewart CR, Monks BG, Reinheckel T, Fitzgerald KA, Latz E, Moore KJ, Golenbock DT. The NALP3 inflammasome is involved in the innate immune response to amyloid-β. Nature Immunology, 2008, 9(8): 857-865. DOI:10.1038/ni.1636 |
| [15] | Hornung V, Bauernfeind F, Halle A, Samstad EO, Kono H, Rock KL, Fitzgerald KA, Latz E. Silica crystals and aluminum salts activate the NALP3 inflammasome through phagosomal destabilization. Nature Immunology, 2008, 9(8): 847-856. DOI:10.1038/ni.1631 |
| [16] | Hassane M, Demon D, Soulard D, Fontaine J, Keller LE, Patin EC, Porte R, Prinz I, Ryffel B, Kadioglu A, Veening JW, Sirard JC, Faveeuw C, Lamkanfi M, Trottein F, Paget C. Neutrophilic NLRP3 inflammasome-dependent IL-1β secretion regulates the γδT17 cell response in respiratory bacterial infections. Mucosal Immunology, 2017, 10(4): 1056-1068. DOI:10.1038/mi.2016.113 |
| [17] | Shoma S, Tsuchiya K, Kawamura I, Nomura T, Hara H, Uchiyama R, Daim S, Mitsuyama M. Critical involvement of pneumolysin in production of interleukin-1alpha and caspase-1-dependent cytokines in infection with Streptococcus pneumoniae in vitro:a novel function of pneumolysin in caspase-1 activation. Infection and Immunity, 2008, 76(4): 1547-1557. DOI:10.1128/IAI.01269-07 |
| [18] | Zhang TJ, Du HH, Feng SW, Wu R, Chen TT, Jiang JL, Peng YY, Ye C, Fang RD. NLRP3/ASC/Caspase-1 axis and serine protease activity are involved in neutrophil IL-1β processing during Streptococcus pneumoniae infection. Biochemical and Biophysical Research Communications, 2019, 513(3): 675-680. DOI:10.1016/j.bbrc.2019.04.004 |
| [19] | Gupta R, Ghosh S, Monks B, DeOliveira RB, Tzeng TC, Kalantari P, Nandy A, Bhattacharjee B, Chan J, Ferreira F, Rathinam V, Sharma S, Lien E, Silverman N, Fitzgerald K, Firon A, Trieu-Cuot P, Henneke P, Golenbock DT. RNA and β-hemolysin of group B Streptococcus induce interleukin-1β(IL-1β) by activating NLRP3 inflammasomes in mouse macrophages. Journal of Biological Chemistry, 2014, 289(20): 13701-13705. DOI:10.1074/jbc.C114.548982 |
| [20] | Fang RD, Tsuchiya K, Kawamura I, Shen YN, Hara H, Sakai S, Yamamoto T, Fernandes-Alnemri T, Yang RL, Hernandez-Cuellar E, Dewamitta SR, Xu YT, Qu HX, Alnemri ES, Mitsuyama M. Critical roles of ASC inflammasomes in caspase-1 activation and host innate resistance to Streptococcus pneumoniae infection. Journal of Immunology, 2011, 187(9): 4890-4899. DOI:10.4049/jimmunol.1100381 |
| [21] | Rabes A, Suttorp N, Opitz B. Inflammasomes in pneumococcal infection: Innate immune sensing and bacterial evasion strategies//Backert S. Inflammasome Signaling and Bacterial Infections. Cham: Springer, 2016. |
| [22] | Zhao WT, Pan F, Wang BJ, Wang C, Sun Y, Zhang TD, Shi YY, Zhang H. Epidemiology characteristics of Streptococcus pneumoniae from children with pneumonia in Shanghai:A retrospective study. Frontiers in Cellular and Infection Microbiology, 2019, 9: 258. DOI:10.3389/fcimb.2019.00258 |
| [23] | Koppe U, Högner K, Doehn JM, Müller HC, Witzenrath M, Gutbier B, Bauer S, Pribyl T, Hammerschmidt S, Lohmeyer J, Suttorp N, Herold S, Opitz B. Streptococcus pneumoniae stimulates a STING-and IFN regulatory factor 3-dependent type I IFN production in macrophages, which regulates RANTES production in macrophages, cocultured alveolar epithelial cells, and mouse lungs. Journal of Immunology, 2012, 188(2): 811-817. DOI:10.4049/jimmunol.1004143 |
| [24] | Karthikeyan RS, Priya JL, Leal SM Jr, Toska J, Rietsch A, Prajna V, Pearlman E, Lalitha P. Host response and bacterial virulence factor expression in Pseudomonas aeruginosa and Streptococcus pneumoniae corneal ulcers. PLoS One, 2013, 8(6): e64867. DOI:10.1371/journal.pone.0064867 |
| [25] | Hara H, Seregin SS, Yang DH, Fukase K, Chamaillard M, Alnemri ES, Inohara N, Chen GY, Núñez G. The NLRP6 inflammasome recognizes lipoteichoic acid and regulates gram-positive pathogen infection. Cell, 2018, 175(6): 1651-1664. DOI:10.1016/j.cell.2018.09.047 |
| [26] | McNeela EA, Burke A, Neill DR, Baxter C, Fernandes VE, Ferreira D, Smeaton S, El-Rachkidy R, McLoughlin RM, Mori A, Moran B, Fitzgerald KA, Tschopp J, Pétrilli V, Andrew PW, Kadioglu A, Lavelle EC. Pneumolysin activates the NLRP3 inflammasome and promotes proinflammatory cytokines independently of TLR4. PLoS Pathogens, 2010, 6(11): e1001191. DOI:10.1371/journal.ppat.1001191 |
| [27] | Karmakar M, Katsnelson M, Malak HA, Greene NG, Howell SJ, Hise AG, Camilli A, Kadioglu A, Dubyak GR, Pearlman E. Neutrophil IL-1β processing induced by pneumolysin is mediated by the NLRP3/ASC inflammasome and caspase-1 activation and is dependent on K+ efflux. Journal of Immunology, 2015, 194(4): 1763-1775. DOI:10.4049/jimmunol.1401624 |
| [28] | Koedel U, Frankenberg T, Kirschnek S, Obermaier B, Hacker H, Paul R, Häcker G. Apoptosis is essential for neutrophil functional shutdown and determines tissue damage in experimental pneumococcal meningitis. PLoS Pathogens, 2009, 5(5): e1000461. DOI:10.1371/journal.ppat.1000461 |
| [29] | Witzenrath M, Pache F, Lorenz D, Koppe U, Gutbier B, Tabeling C, Reppe K, Meixenberger K, Dorhoi A, Ma JT, Holmes A, Trendelenburg G, Heimesaat MM, Bereswill S, van der Linden M, Tschopp J, Mitchell TJ, Suttorp N, Opitz B. The NLRP3 inflammasome is differentially activated by pneumolysin variants and contributes to host defense in pneumococcal pneumonia. Journal of Immunology, 2011, 187(1): 434-440. DOI:10.4049/jimmunol.1003143 |
| [30] | Muñoz-Planillo R, Kuffa P, Martínez-Colón G, Smith BL, Rajendiran TM, Núñez G. K+ efflux is the common trigger of NLRP3 inflammasome activation by bacterial toxins and particulate matter. Immunity, 2013, 38(6): 1142-1153. DOI:10.1016/j.immuni.2013.05.016 |
| [31] | Feng SW, Huang QY, Ye C, Wu R, Lei GH, Jiang JL, Chen TT, Peng YY, Fang RD. Syk and JNK signaling pathways are involved in inflammasome activation in macrophages infected with Streptococcus pneumoniae. Biochemical and Biophysical Research Communications, 2018, 507(1/4): 217-222. |
| [32] | Moon JS, Nakahira K, Chung KP, DeNicola GM, Koo MJ, Pabón MA, Rooney KT, Yoon JH, Ryter SW, Stout-Delgado H, Choi AMK. NOX4-dependent fatty acid oxidation promotes NLRP3 inflammasome activation in macrophages. Nature Medicine, 2016, 22(9): 1002-1012. DOI:10.1038/nm.4153 |
| [33] | Parker D. Impact of type I and III interferons on respiratory superinfections due to multidrug-resistant pathogens. Journal of Infectious Diseases, 2017, 215(S1): S58-S63. |
| [34] | Weigent DA, Huff TL, Peterson JW, Stanton GJ, Baron S. Role of interferon in streptococcal infection in the mouse. Microbial Pathogenesis, 1986, 1(4): 399-407. DOI:10.1016/0882-4010(86)90071-9 |
| [35] | Fang RD, Hara H, Sakai S, Hernandez-Cuellar E, Mitsuyama M, Kawamura I, Tsuchiya K. Type I interferon signaling regulates activation of the absent in melanoma 2 inflammasome during Streptococcus pneumoniae infection. Infection and Immunity, 2014, 82(6): 2310-2317. DOI:10.1128/IAI.01572-14 |
| [36] | Dela Cruz CS, Liu W, He CH, Jacoby A, Gornitzky A, Ma B, Flavell R, Lee CG, Elias JA. Chitinase 3-like-1 promotes Streptococcus pneumoniae killing and augments host tolerance to lung antibacterial responses. Cell Host & Microbe, 2012, 12(1): 34-46. |
| [37] | Erttmann SF, Gekara NO. Hydrogen peroxide release by bacteria suppresses inflammasome-dependent innate immunity. Nature Communications, 2019, 10(1): 3493. DOI:10.1038/s41467-019-11169-x |
| [38] | Bauernfeind F, Bartok E, Rieger A, Franchi L, Núñez G, Hornung V. Cutting edge:reactive oxygen species inhibitors block priming, but not activation, of the NLRP3 inflammasome. Journal of Immunology, 2011, 187(2): 613-617. DOI:10.4049/jimmunol.1100613 |
| [39] | Feng SW, Chen TT, Lei GH, Hou FQ, Jiang JL, Huang QY, Peng YY, Ye C, Hu DL, Fang RD. Absent in melanoma 2 inflammasome is required for host defence against Streptococcus pneumoniae infection. Innate Immunity, 2019, 25(7): 412-419. DOI:10.1177/1753425919860252 |
| [40] | Hoegen T, Tremel N, Klein M, Angele B, Wagner H, Kirschning C, Pfister HW, Fontana A, Hammerschmidt S, Koedel U. The NLRP3 inflammasome contributes to brain injury in pneumococcal meningitis and is activated through ATP-dependent lysosomal cathepsin B release. Journal of Immunology, 2011, 187(10): 5440-5451. DOI:10.4049/jimmunol.1100790 |
| [41] | Zwijnenburg PJ, van der Poll T, Florquin S, Akira S, Takeda K, Roord JJ, van Furth AM. Interleukin-18 gene-deficient mice show enhanced defense and reduced inflammation during pneumococcal meningitis. Journal of Neuroimmunology, 2003, 138(1/2): 31-37. |
| [42] | Geldhoff M, Mook-Kanamori BB, Brouwer MC, Troost D, Leemans JC, Flavell RA, Van der Ende A, Van der Poll T, Van de Beek D. Inflammasome activation mediates inflammation and outcome in humans and mice with pneumococcal meningitis. BMC Infectious Diseases, 2013, 13(1): 358. |
| [43] | Mitchell AJ, Yau B, McQuillan JA, Ball HJ, Too LK, Abtin A, Hertzog P, Leib SL, Jones CA, Gerega SK, Weninger W, Hunt NH. Inflammasome-dependent IFN-γ drives pathogenesis in Streptococcus pneumoniae meningitis. Journal of Immunology, 2012, 189(10): 4970-4980. DOI:10.4049/jimmunol.1201687 |
| [44] | Lauw FN, Branger J, Florquin S, Speelman P, van Deventer SJ, Akira S, van der Poll T. IL-18 improves the early antimicrobial host response to pneumococcal pneumonia. Journal of Immunology, 2002, 168(1): 372-378. DOI:10.4049/jimmunol.168.1.372 |
| [45] | Lemon JK, Miller MR, Weiser JN. Sensing of interleukin-1 cytokines during Streptococcus pneumoniae colonization contributes to macrophage recruitment and bacterial clearance. Infection and Immunity, 2015, 83(8): 3204-3212. DOI:10.1128/IAI.00224-15 |
| [46] | Kostadinova E, Chaput C, Gutbier B, Lippmann J, Sander LE, Mitchell TJ, Suttorp N, Witzenrath M, Opitz B. NLRP3 protects alveolar barrier integrity by an inflammasome-independent increase of epithelial cell adherence. Scientific Reports, 2016, 6(1): 30943. DOI:10.1038/srep30943 |
| [47] | Fang RD, Uchiyama R, Sakai S, Hara H, Tsutsui H, Suda T, Mitsuyama M, Kawamura I, Tsuchiya K. ASC and NLRP3 maintain innate immune homeostasis in the airway through an inflammasome-independent mechanism. Mucosal Immunology, 2019, 12(5): 1092-1103. DOI:10.1038/s41385-019-0181-1 |
| [48] | Littmann M, Albiger B, Frentzen A, Normark S, Henriques-Normark B, Plant L. Streptococcus pneumoniae evades human dendritic cell surveillance by pneumolysin expression. EMBO Molecular Medicine, 2009, 1(4): 211-222. DOI:10.1002/emmm.200900025 |
| [49] | Mitchell TJ, Mendez F, Paton JC, Andrew PW, Boulnois GJ. Comparison of pneumolysin genes and proteins from Streptococcus pneumoniae types 1 and 2. Nucleic Acids Research, 1990, 18(13): 4010. DOI:10.1093/nar/18.13.4010 |
| [50] | Hausdorff WP, Feikin DR, Klugman KP. Epidemiological differences among pneumococcal serotypes. Lancet Infectious Diseases, 2005, 5(2): 83-93. |
| [51] | Kirkham LAS, Jefferies JM, Kerr AR, Yu J, Clarke SC, Andrew S, Mitchell TJ. Identification of invasive serotype 1 pneumococcal isolates that express nonhemolytic pneumolysin. Journal of Clinical Microbiology, 2006, 44(1): 151-159. DOI:10.1128/JCM.44.1.151-159.2006 |
| [52] | Jefferies JMC, Johnston CHG, Kirkham LAS, Cowan GJM, Ross KS, Smith A, Clarke SC, Brueggemann AB, George RC, Pichon B, Pluschke G, Pfluger V, Mitchell TJ. Presence of nonhemolytic pneumolysin in serotypes of Streptococcus pneumoniae associated with disease outbreaks. Journal of Infectious Diseases, 2007, 196(6): 936-944. DOI:10.1086/520091 |
| [53] | Fatykhova D, Rabes A, Machnik C, Guruprasad K, Pache F, Berg J, Toennies M, Bauer TT, Schneider P, Schimek M, Eggeling S, Mitchell TJ, Mitchell AM, Hilker R, Hain T, Suttorp N, Hippenstiel S, Hocke AC, Opitz B. Serotype 1 and 8 pneumococci evade sensing by inflammasomes in human lung tissue. PLoS One, 2015, 10(8): e0137108. DOI:10.1371/journal.pone.0137108 |
| [54] | Weinberger DM, Harboe ZB, Sanders EAM, Ndiritu M, Klugman KP, Rückinger S, Dagan R, Adegbola R, Cutts F, Johnson HL, O'Brien KL. Association of serotype with risk of death due to pneumococcal pneumonia:a meta-analysis. Clinical Infectious Diseases, 2010, 51(6): 692-699. DOI:10.1086/655828 |
| [55] | Le Hello S, Watson M, Levy M, Marcon S, Brown M, Yvon JF, Missotte I, Garin B. Invasive serotype 1Streptococcus pneumoniae outbreaks in the South Pacific from 2000 to 2007. Journal of Clinical Microbiology, 2010, 48(8): 2968-2971. DOI:10.1128/JCM.01615-09 |
| [56] | Sandgren A, Sjöström K, Olsson Liljequist B, Christensson B, Samuelsson A, Kronvall G, Henriques NB. Effect of clonal and serotype-specific properties on the invasive capacity of Streptococcus pneumoniae. Journal of Infectious Diseases, 2004, 189(5): 785-796. DOI:10.1086/381686 |
| [57] | Davis KM, Nakamura S, Weiser JN. Nod2 sensing of lysozyme-digested peptidoglycan promotes macrophage recruitment and clearance of S. pneumoniae colonization in mice. Journal of Clinical Investigation, 2011, 121(9): 3666-3676. DOI:10.1172/JCI57761 |
| [58] | Cole J, Aberdein J, Jubrail J, Dockrell DH. The role of macrophages in the innate immune response to Streptococcus pneumoniae and Staphylococcus aureus:mechanisms and contrasts. Advances in Microbial Physiology, 2014, 65: 125-202. DOI:10.1016/bs.ampbs.2014.08.004 |
| [59] | Maltez VI, Miao EA. Reassessing the evolutionary importance of inflammasomes. Journal of Immunology, 2016, 196(3): 956-962. DOI:10.4049/jimmunol.1502060 |
 2020, Vol. 60
2020, Vol. 60