中国科学院微生物研究所,中国微生物学会,中国菌物学会
文章信息
- Wanqian Yan, Xinyuan Mao, Yujun Dai, Lihua Wang, Cuiying Du, Cao Zheng. 2021
- 严婉芊, 毛馨缘, 戴余军, 王立华, 都萃颖, 郑操. 2021
- Enzymatic characterization of TbrA, the first identified aconitate isomerase from Bacillus thuringiensis
- 分离自苏云金芽胞杆菌的首例细菌乌头酸异构酶TbrA的酶学性质研究
- Acta Microbiologica Sinica, 61(2): 388-397
- 微生物学报, 61(2): 388-397
-
文章历史
- 收稿日期:2020-03-22
- 修回日期:2020-06-09
- 网络出版日期:2020-08-17
2. Hubei Key Laboratory of Quality Control of Characteristic Fruits and Vegetables, Hubei Engineering University, Xiaogan 432000, Hubei Province, China
2. 湖北工程学院特色果蔬质量安全控制湖北省重点实验室, 湖北 孝感 432000
Aconitate isomerase (AI; EC 5.3.3.7) catalyzes the interconversion between cis-aconitic acid (CAA) and trans-aconitic acid (TAA)[1–3], a metabolic branch of the central tricarboxylic acid (TCA) cycle (Figure 1). Rao and Altekar first reported the existence of AI in 1961 in some Pseudomonas bacteria, that could grow on a synthetic medium with TAA as the sole carbon source (later called the ACO medium[4–5]); they observed that their cell-free extracts could mediate the formation of citric acid from TAA as well as TAA from CAA in vitro[6]. In addition, many plants such as maize, wheat, barnyard grass and sugar cane were also found to produce AI[7–9], which generally exists at high concentrations in the leaf and root tissues[7]. Although its existence has been proposed for more than half a century, the first AI-encoding gene, tbrA (TAA biosynthesis-related gene A), was just identified recently in our research on Bacillus thuringiensis, one of the most excellent nematode pathogens[10–13], by both in vivo and in vitro experiments[1]. However, AI genes of plant origin continue to remain unknown. The tbrA gene (1074 bp) is located on the B. thuringiensis plasmid pCT281 and encodes TbrA (357 aa), a cytoplasmic protein that belongs to the PrpF isomerase superfamily. The sequence of TbrA shares 27% identify and 97% coverage with that of PrpF (397 aa) in Shewanella oneidensis strain MR-1[1, 14–15]. Another gene, tbrB, which is located 111 bp downstream of tbrA, encodes a TAA extracellular-transporter protein and comprises a tbr operon with tbrA. However, the physiological function mediated by TbrA is different from the function of TAA assimilation mediated by most of the other AI proteins[4, 6], TbrA is a seemingly new and distinct type of AI protein because it cannot support the growth of B. thuringiensis on an ACO medium[16], however, it could specifically enable the cell to produce considerable amounts of TAA. The significance of high TAA production was later demonstrated as that TAA is a natural nematicidal factor, which is specifically biosynthesized by the B. thuringiensis pathogen to intoxicate nematode hosts[1]. These findings revealed the first biological sequence of AI protein, and may facilitate the progress of identifying the AI gene in plant species.

|
| Figure 1 Aconitate isomerase mediates a specific branch of the TCA cycle. A: Chemical structural formula of TAA; B: Chemical structural formula of CAA. The conformational differences between the 1-carboxyl groups of TAA and CAA molecules are highlighted in blue and red, respectively; C: The relationship between the AI-mediated pathway and the TCA cycle. |
TAA has the potential for application in many fields. On one hand, TAA has long been used as industrial chemicals like antioxidant, plasticizer, and lubricant and as a substrate of functional polymers[17–19]. On the other hand, TAA shows attractive potential for applications in agriculture and medicine. In addition to intoxicate nematodes[16], for example killing the most damaging root-knot and cyst nematodes[20–22], TAA is also active against the brown planthopper (the most notorious pest of rice)[8, 23–26], the Leishmania parasite (the pathogen of kala-azar disease)[27–28], and mammalian inflammation[29–30]. However, presently, TAA is mainly produced through industrial chemosynthesis[31], a process that is accompanied by the release of numerous byproducts. Hence, an effective, selective, and eco-friendly enzymatic bioprocess needs to be developed for TAA production, and one possible improvement includes inclusion of a TbrA-based biocatalyst. Therefore, the catalytic properties of this functional enzyme should be elucidated.
In this study, the enzymatic characteristics of TbrA, the sole AI protein that is specific in TAA biosynthesis, were quantitatively determined under optimal reaction conditions including cofactor effects, optimal pH, temperature as well as ionic strength parameters, and kinetic constants of Km, Vmax, kcat, and kcat/Km. This work provides a more detailed understanding of AI catalysis and may serve as a foundation for utilizing enzymatic engineering for the industrial bioproduction of TAA in the future.
1 Materials and methods 1.1 Bacterial strains and culture conditionsThe tbrA gene (1074 bp) is located on the largest plasmid, namely the pCT281 (281231 bp) plasmid of the B. thuringiensis CT-43 strain[32]. Thus, the CT-43 strain was used as a tbrA gene donor. Escherichia coli DH5α and Rosetta strains were used for tbrA cloning and TbrA expression, respectively. All B. thuringiensis and E. coli strains were cultured in Luria-Bertani (LB) medium at 28 ℃ for both CT-43 and Rosetta cells, and at 37 ℃ for DH5α cells. When appropriate, antibiotics were added at the following final concentrations: 100 μg/mL ampicillin, 25 μg/mL chloramphenicol, and 50 μg/mL kanamycin.
1.2 Construction of a TbrA-expression plasmid and transformation of bacterial cellsDNA fragment harboring the open reading frame (ORF) region of tbrA (stop codon-excluded; 1071 bp) was amplified from the genomic DNA of B. thuringiensis CT-43 strain and cloned into Nde I and Xho I sites of the pET28a expression vector to generate a recombinant pET28a-tbrA plasmid. Thereafter, the plasmid was transformed firstly into E. coli DH5α cells for sequence verification, and finally into E. coli Rosetta cells, to yield recombinant Rosetta-TbrA for subsequent TbrA-inducible expression. The primer sequences for tbrA ORF amplification were as follows: tbrA-F-Nde I, GG AATTCCATATGATGAAAATACCTTGTTTTGTT, and tbrA-R-Xho I, GCCCTCGAGAGGTATTATT AATTCGCCTTT. Nde I and Xho I restriction sites are underlined.
1.3 Purification of recombinant TbrAA single colony of Rosetta-TbrA cells was cultured in 5 mL of LB medium with kanamycin and chloramphenicol at 37 ℃ for 4 h. Subsequently, the culture was transferred into 100 mL of LB medium with an appropriate amount of antibiotics in a ratio of 1:100, and was cultured at 37 ℃ to an OD600 of 0.8. Following the addition of IPTG (final concentration of 0.1 mmol/L), the TbrA protein was induced at 28 ℃ for 6 h with vigorous shaking. Cells were harvested and subjected to high-pressure homogenization at 4 ℃. Thereafter, the generated cell-free extract was loaded onto a Ni2+ nitrilotriacetic acid column, on which the C-terminal His6-tagged TbrA was remained. The column was washed and the His-tagged TbrA was eluted using imidazole at concentrations of 60 and 500 mmol/L, respectively. The collected fractions were dialyzed against 20 mmol/L Tris-HCl (pH 8.0), 10% glycerol, 1 mmol/L EDTA, 0.1 mmol/L DTT, and subsequently stored at –80 ℃.
1.4 HPLC quantification of CAA and TAA productionThe HPLC system used for determining the amount of CAA and TAA synthesized in the enzymatic reaction systems was composed of a HITACHI Primaide 1110 pump, a HITACHI Primaide 1430 diode array detector, and a TC-C18 column (250 mm×4.6 mm, 5 μm; Agilent). To quantify the CAA content, a standard curve of CAA concentration against the CAA peak area was first plotted and the external standard method was used. Thereafter, a 10-μL volume of reaction sample was delivered at a flow rate of 1.0 mL/min by the mobile phase containing 10% methanol with 0.1% formic acid, and was monitored at 260 nm and at 25 ℃ for 15 min. The CAA and TAA peaks were efficiently separated under the conditions described above. The TAA content was quantified by adopting the same aforementioned procedure; however, a standard curve with TAA concentration against TAA peak area was plotted. CAA and TAA commercial standards were purchased from Sigma-Aldrich and Tokyo Chemical Industry, respectively.
1.5 Characterization of the AI activity of TbrA1.5.1 Influence of pH and temperature on enzyme activity: All enzymatic activity assays were performed using 3.4 μmol/L purified TbrA (to a final concentration of 0.34 μmol/L), which was dissolved in the dialysate buffer containing 20 mmol/L Tris-HCl (pH 8.0), 10% glycerol, 1 mmol/L EDTA, and 0.1 mmol/L DTT. The TbrA enzymatic assay was performed by firstly adjusting the pH of CAA or TAA substrate samples to 7.0 by adding 5 mol/L NaOH prior to their addition to the reaction system. Along with each of the characterization assays, a control reaction, in which the purified TbrA enzyme was excluded, was also assayed, so that the non-enzymatic isomerization between CAA and TAA could be deducted from the total product formation of each reaction. All reactions were conducted at 250 μL test volume at a relevant designated temperature for 30 min. The reactions were terminated by the addition of 10 μL of 6 mol/L HCl, and the samples were subsequently analyzed by HPLC as mentioned before. In this study, all experiments were preformed in triplicates, and the error bars represented the standard deviation of the three replicates.
The optimum pH of TbrA catalysis was determined by performing an enzymatic assay using different buffers (as described in the legend of Figure 2) at 37 ℃. Additionally, the optimal temperature was determined by incubating each of the samples at various temperatures including 10, 20, 28, 37, 50, 60, 70 ℃ at pH 8.0.

|
| Figure 2 Biochemical characterization of TbrA at various pH levels and temperatures. A: SDS-PAGE of the purified TbrA. To purify TbrA, the recombinant protein with a His6-tag was overexpressed in E. coli Rosetta-TbrA cells and purified using Ni2+ affinity chromatography and dialysis. The protein marker size has been indicated. B: The effect of pH on TbrA activity. The activity of TbrA was measured at 37 ℃ under various pH conditions: black squares, 5.5–6.5 (50 mmol/L MES); red circles, 7.0–8.0 (50 mmol/L HEPES); blue triangles, 8.5–10.0 (50 mmol/L CHES). C: The effect of temperature on TbrA activity. The enzyme activity of TbrA was measured at temperatures ranging from 10 ℃ to 70 ℃ in 50 mmol/L HEPES buffer (pH 8.0). |
1.5.2 Effect of metal ions and DTT: To evaluate the effect of metal ions and DTT on the isomeric activity of TbrA, the enzymatic systems containing CAA as the substrate were treated with a 10 mmol/L final concentration of MgCl2, MnCl2, CaCl2, ZnCl2, FeCl2, CuCl2, NiSO4, CoCl2, and DTT solution in 50 mmol/L HEPES buffer (pH 8.0), at 37 ℃ for 30 min. TbrA activity in the control tube that contained no additives was set to 100%; thereafter, the relative activity of each additive-containing tube was calculated. To examine the optimal concentration of metal ion that has the maximum stimulation effect on enzymatic activity, the ion samples with final concentrations ranging from 10 mmol/L to 250 mmol/L were used.
1.5.3 Effect of ionic strength: The effect of ionic strength on the activity of TbrA was evaluated by incubating the reaction supernatant in 50 mmol/L HEPES buffer (pH 8.0), containing NaCl at concentrations ranging from 0 to 500 mmol/L, at 37 ℃ for 30 min. The highest isomeric activity was set to 100%.
1.5.4 Kinetic parameters of TbrA enzyme: The Km and Vmax values of TbrA were measured in reaction samples that comprised of varying final concentrations of CAA (1, 2, 5, 10, 20, 50 mmol/L) or TAA (1, 2.5, 5.0, 10.0, 25.0, 50.0 mmol/L) in 50 mmol/L HEPES buffer (pH 8.0) at 37 ℃ for 30 min. The Km and Vmax kinetic parameters were determined using the Lineweaver-Burk plot. The concentration of TbrA enzyme used for kcat and kcat/Km calculation was determined by the Bradford method[33], for which bovine serum albumin was used as the standard.
2 Results and discussion 2.1 Cloning, expression, and purification of TbrAThe DNA fragment of tbrA ORF (1071 bp, the stop-codon 'TAA' was excluded), which was amplified from the genomic DNA of the B. thuringiensis CT-43 strain, was cloned into the pET28a expression vector and introduced into E. coli Rosetta cells for TbrA-inducible expression. After inducing the cells with 0.1 mmol/L IPTG for 6 h at 28 ℃, the C-terminal His6-tagged TbrA was released into the cell supernatant by a high-pressure homogenizer, and subsequently purified using Ni2+ affinity chromatography. SDS-PAGE analysis of the eluted fraction revealed a band of approximately 40 kDa (Figure 2-A), which corresponds to the theoretical molecular weight of the His6-tagged TbrA (41487 Da), as predicted by the Compute pI/Mw tool available from ExPASy. Consequently, about 1 mg of TbrA protein was obtained from 200 mL of cell culture in LB medium.
2.2 Effect of pH and temperature on TbrA activityGenerally, AI catalyzes both the forward (from CAA to TAA) and the reverse (from TAA to CAA) isomerization reactions. In this study, we chose the forward reaction, which is responsible for TAA biosynthesis, to determine the optimal conditions for TbrA catalysis.
To detect the effect of pH, TbrA activity was measured at pH values ranging from 5.5 to 10.0, using 50 mmol/L MES buffer (pH 5.5–6.5), 50 mmol/L HEPES buffer (pH 7.0–8.0), and 50 mmol/L CHES buffer (pH 8.5–10.0). The reactions were performed at 37 ℃ for 30 min using 10 mmol/L CAA as the substrate. As shown in Figure 2-B, TbrA could efficiently synthesize TAA in weakly alkaline conditions (pH 7.5–8.0); the highest relative TbrA activity was observed at pH 8.0, but which rapidly decreased at the strongly alkaline pH range. Therefore, unless otherwise specified, 50 mmol/L HEPES at pH 8.0 was used for TbrA reactions.
The effect of temperature was analyzed from 10 to 70 ℃. The optimal temperature for TbrA activity was observed to be 37 ℃ (Figure 2-C). Clearly, TbrA has a relatively wide suitable temperature range and is more tolerant to low temperatures, as nearly 60% of the relative activity was stored at 10 ℃, whereas at 70 ℃, only 30% of the activity remained. This characteristic may allow for higher product output with lower industrial energy input, which may be advantageous for possible future industrial applications of TbrA. Therefore, the reaction temperature was controlled at 37 ℃ in the following characterization experiments.
2.3 Effect of chemical additives on TbrA activityThe effect of eight metal ions and DTT chemical on TbrA activity was assessed. As shown in Figure 3-A, Mg2+, followed by Ca2+, exhibits the strongest activating effect. Figure 3-B indicates that the optimal Mg2+ concentration is 75 mmol/L. Furthermore, DTT did not inhibit, but rather promoted TbrA activity (Figure 3-A), suggesting the absence of disulfide bonds in TbrA, despite the presence of five Cys residues in its amino acid sequence. On the contrary, most divalent metal cations including Mn2+, Zn2+, Cu2+, Fe2+, Co2+ and Ni2+ inhibited TbrA activity to different extents, thus, indicating that chemical contamination of these metal cations should be avoided in TbrA applications. A previous study showed that an AI protein, which was non-specifically isolated from the cell-free extract of the Pseudomonas putida A 3.12 strain, required Fe2+ for its activity[2]. In contrast, our finding shows TbrA activity to be independent of any metal cofactors, with Fe2+ even showing a significant inhibitory effect (Figure 3-A). It is possible that AI from different bacterial species evolved independently and specifically, and thus exhibits distinct cofactor preferences. However, this speculation requires confirmation through more efforts in isolation of AI enzymes from different bacterial species in the future.
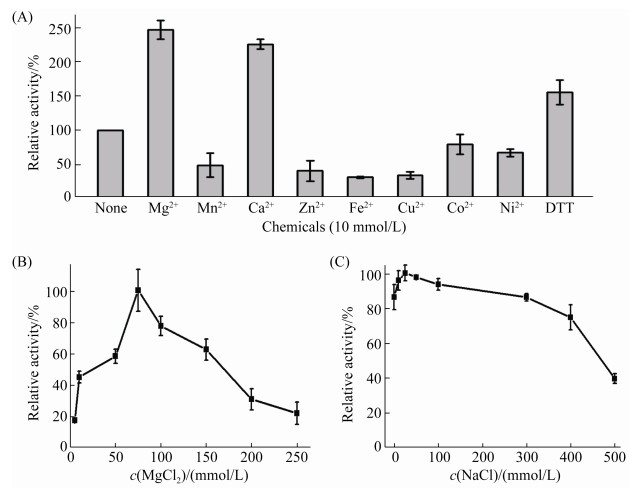
|
| Figure 3 Biochemical characterization of TbrA regarding inclusion of chemical additives and ionic strength. A: The effect of metal ions and DTT on TbrA activity. This effect was determined at 37 ℃ in 50 mmol/L HEPES buffer (pH 8.0), containing 10 mmol/L final concentration of either DTT or various metal ions (MgCl2, MnCl2, CaCl2, ZnCl2, FeCl2, CuCl2, NiSO4, CoCl2). B: The optimal concentration of Mg2+ on TbrA activity. The activity of TbrA was measured at 37 ℃ under various final concentrations of MgCl2, ranging from 10 to 250 mmol/L in 50 mmol/L HEPES buffer (pH 8.0). C: The effect of ionic strength on TbrA activity. The enzyme activity of TbrA was measured at 37 ℃ under various final concentrations of NaCl, ranging from 0 to 500 mmol/L, in 50 mmol/L HEPES buffer (pH 8.0). |
2.4 Effect of ionic strength on TbrA activity
A series of tests were carried out by adding increasing amount of NaCl to the reaction samples (0–500 mmol/L). The activity of TbrA was found to be initially activated at low NaCl concentration, but attained maximum activity at 25 mmol/L; thereafter, TbrA activity was inversely correlated to the increase in NaCl concentration, ranging between 100 mmol/L to 500 mmol/L. This result suggests that maintaining appropriate ionic strength would be beneficial for TbrA reaction.
2.5 Kinetic characterization of the AI protein TbrAThe Km, Vmax, kcat, and kcat/Km of TbrA enzyme for both its forward and reverse reactions were calculated (Figure 4) and summarized in Table 1. It can be concluded that although TbrA mediates the interconversion between aconitic acid isomers, the affinity (Km) and the catalytic efficiency (kcat/Km) of TbrA were much higher when CAA, rather than TAA, was used as the substrate, suggesting that TbrA favors the equilibrium for TAA formation. This is consistent with the conventional knowledge of AI protein[2]. Hence, minimizing CAA formation, resulting from the reverse reaction, by directed mutagenesis of tbrA may be worth considering to increase the TAA yield.

|
| Figure 4 Lineweaver-Burk plots for determining the kinetic parameters of TbrA. A: Forward reaction with CAA as the substrate; B: Reverse reaction with TAA as the substrate. Data are expressed as the mean of triplicates. |
| Substrates | Km/ (mmol/L) |
Vmax/ (μmol/L·s) |
kcat/(1/s) | (kcat/Km)/ (L/mmol·s) |
| CAA | 6.25 | 1.39 | 4.08 | 0.65 |
| TAA | 71.50 | 4.17 | 12.25 | 0.17 |
Consistent with the increased CAA consumption to form TAA that was observed in vitro, TbrA also specifically contributes to the net production of TAA in B. thuringiensis cells in vivo. Notably, the majority of reported AI proteins (primarily in Pseudomonas spp.) display similar preference for TAA formation as TbrA; however, they specifically mediate the net consumption and not the net production of TAA[4, 6]. The reason behind the distinct physiological functions of similarly-behaved AI proteins has not been studied thus far. Here, we proposed two hypotheses. First, the intracellular biochemical environments of B. thuringiensis and Pseudomonas are different and comprise some distinct active substance(s), which favor opposite directions of AI catalysis, thus resulting in distinct physiological functions. Second, the bacterial cells may evolve two types of TAA transporters, one for intracellular-transport and the other for extracellular-transport, and these two functions are highly independent. Notably, in B. thuringiensis CT-43 strain, we found the TAA extracellular transporter TbrB to be essential for TAA production[1]. Therefore, deletion of tbrB abolished TAA biosynthesis to prevent the accumulation of TAA molecules so that its toxic effect on the important enzymes involved in central metabolisms can be reduced[34–35]. Meanwhile, with the presence of TbrA, the CT-43 cell could not assimilate TAA, indicating that although anchored on the cell surface, TbrB cannot transport TAA intracellularly. As for the TAA-assimilating bacteria, they may employ an intracellular transporter to avail TAA as a substrate for the AI enzymes, so that the carbon source can be incorporated into the TCA cycle through the CAA metabolite. Whether the TAA-assimilating bacteria are further able to produce TAA depends on their ability to encode the extracellular TAA transporter. To date, no relevant gene or protein sequence has been reported; however, our group is currently conducting research to identify extracellular transporter genes in TAA-assimilating bacteria.
3 ConclusionThe aconitate isomerase TbrA from B. thuringiensis prefers CAA as substrate to catalyze TAA production. The highest enzymatic activity of TbrA was achieved at moderate pH and temperature conditions and was enhanced by 75 mmol/L Mg2+ and 25 mmol/L NaCl. The biosafe and multi-active TAA molecule has potential for use in crop pest control, with wider application in the polymer industry and even disease treatment in the future. This work is the first to determine the catalytic characteristics of the TAA-biosynthesizing enzyme and may serve as the basis for future use of the enzyme in the industrial bioproduction of TAA.
| [1] | Du CY, Cao SY, Shi XY, Nie XT, Zheng JS, Deng Y, Ruan LF, Peng DH, Sun M. Genetic and biochemical characterization of a gene operon for trans-aconitic acid, a novel nematicide from Bacillus thuringiensis. The Journal of Biological Chemistry, 2017, 292(8): 3517-3530. DOI:10.1074/jbc.M116.762666 |
| [2] | Klinman JP, Rose IA. Purification and kinetic properties of aconitate isomerase from Pseudomonas putida. Biochemistry, 1971, 10(12): 2253-2259. DOI:10.1021/bi00788a011 |
| [3] | Klinman JP, Rose IA. Mechanism of the aconitate isomerase reaction. Biochemistry, 1971, 10(12): 2259-2266. DOI:10.1021/bi00788a012 |
| [4] | Watanabe K, Katsuhara M, Nakao H, Sato M. Detection and molecular analysis of plantand insect-associated bacteria harboring aconitate isomerase involved in biosynthesis of trans-aconitic acid as antifeedant in brown planthoppers. Current Microbiology, 1997, 35(2): 97-102. DOI:10.1007/s002849900219 |
| [5] |
Zheng C, Cai L, Zhang ZQ, Wang LH, Dai YJ, Du CY. Isolation and identification of soil Bacillus strains that encode aconitate isomerase. Acta Microbiologica Sinica, 2019, 59(7): 1373-1382.
(in Chinese) 郑操, 蔡鹭, 张中强, 王立华, 戴余军, 都萃颖. 土壤中可编码乌头酸异构酶的芽胞杆菌菌株筛选及鉴定. 微生物学报, 2019, 59(7): 1373-1382. |
| [6] | Rao MRR, Altekar WM. Aconitate isomerase. Biochemical and Biophysical Research Communications, 1961, 4(2): 101-105. DOI:10.1016/0006-291X(61)90355-2 |
| [7] | Thompson JF, Schaefer SC, Madison JT. Determination of aconitate isomerase in plants. Analytical Biochemistry, 1990, 184(1): 39-47. DOI:10.1016/0003-2697(90)90008-W |
| [8] | Kim M, Koh HS, Obata T, Fukami H, Ishii S. Isolation and identification of trans-aconitic acid as the antiffeedant in barnyard grass against the brown planthopper, Nilaparvata lugens (STÅL) (Homoptera:Delphacidae). Applied Entomology and Zoology, 1976, 11(1): 53-57. DOI:10.1303/aez.11.53 |
| [9] | Montoya G, Londono J, Cortes P, Izquierdo O. Quantitation of trans-aconitic acid in different stages of the sugar-manufacturing process. Journal of Agricultural and Food Chemistry, 2014, 62(33): 8314-8318. DOI:10.1021/jf5008874 |
| [10] | Bravo A, Likitvivatanavong S, Gill SS, Soberón M. Bacillus thuringiensis:a story of a successful bioinsecticide. Insect Biochemistry and Molecular Biology, 2011, 41(7): 423-431. DOI:10.1016/j.ibmb.2011.02.006 |
| [11] | Ruan LF, Crickmore N, Peng DH, Sun M. Are nematodes a missing link in the confounded ecology of the entomopathogen Bacillus thuringiensis?. Trends in Microbiology, 2015, 23(6): 341-346. DOI:10.1016/j.tim.2015.02.011 |
| [12] | Ruan LF, Wang HH, Cai G, Peng DH, Zhou H, Zheng JS, Zhu L, Wang XX, Yu HQ, Li S, Geng C, Sun M. A two-domain protein triggers heat shock pathway and necrosis pathway both in model plant and nematode. Environmental Microbiology, 2015, 17(11): 4547-4565. DOI:10.1111/1462-2920.12968 |
| [13] | Peng DH, Lin J, Huang Q, Zheng W, Liu GQ, Zheng JS, Zhu L, Sun M. A novel metalloproteinase virulence factor is involved in Bacillus thuringiensis pathogenesis in nematodes and insects. Environmental Microbiology, 2016, 18(3): 846-862. DOI:10.1111/1462-2920.13069 |
| [14] | Grimek TL, Escalante-Semerena JC. The acnD genes of Shewenella oneidensis and Vibrio cholerae encode a new Fe/S-dependent 2-methylcitrate dehydratase enzyme that requires prpF function in vivo. Journal of Bacteriology, 2004, 186(2): 454-462. DOI:10.1128/JB.186.2.454-462.2004 |
| [15] | Garvey GS, Rocco CJ, Escalante-Semerena JC, Rayment I. The three-dimensional crystal structure of the PrpF protein of Shewanella oneidensis complexed with trans-aconitate:insights into its biological function. Protein Science, 2007, 16(7): 1274-1284. DOI:10.1110/ps.072801907 |
| [16] | 都萃颖.小分子杀线虫毒素反式乌头酸生物合成途径及该毒素应用于植物根结线虫防治的研究.华中农业大学博士学位论文, 2017. |
| [17] | Piang-Siong W, De Caro P, Marvilliers A, Chasseray X, Payet B, Shum Cheong Sing A, Illien B. Contribution of trans-aconitic acid to DPPH scavenging ability in different media. Food Chemistry, 2017, 214: 447-452. DOI:10.1016/j.foodchem.2016.07.083 |
| [18] | Cao HL, Zheng Y, Zhou J, Wang WX, Pandit A. A novel hyperbranched polyester made from aconitic acid (B3) and di(ethylene glycol) (A2). Polymer International, 2011, 60(4): 630-634. DOI:10.1002/pi.2993 |
| [19] | Gil N, Saska M, Negulescu I. Evaluation of the effects of biobased plasticizers on the thermal and mechanical properties of poly (vinyl chloride). Journal of Applied Polymer Science, 2006, 102(2): 1366-1373. DOI:10.1002/app.24132 |
| [20] | Jones JT, Haegeman A, Danchin EGJ, Gaur HS, Helder J, Jones MGK, Kikuchi T, Manzanilla-López R, Palomares-Rius JE, Wesemael WML, Perry RN. Top 10 plant-parasitic nematodes in molecular plant pathology. Molecular Plant Pathology, 2013, 14(9): 946-961. DOI:10.1111/mpp.12057 |
| [21] | Abad P, Gouzy J, Aury JM, Castagnone-Sereno P, Danchin EGJ, Deleury E, Perfus-Barbeoch L, Anthouard V, Artiguenave F, Blok VC, Caillaud MC, Coutinho PM, Dasilva C, De Luca F, Deau F, Esquibet M, Flutre T, Goldstone JV, Hamamouch N, Hewezi T, Jaillon O, Jubin C, Leonetti P, Magliano M, Maier TR, Markov GV, McVeigh P, Pesole G, Poulain J, Robinson-Rechavi M, Sallet E, Ségurens B, Steinbach D, Tytgat T, Ugarte E, Van Ghelder C, Veronico P, Baum TJ, Blaxter M, Bleve-Zacheo T, Davis EL, Ewbank JJ, Favery B, Grenier E, Henrissat B, Jones JT, Laudet V, Maule AG, Quesneville H, Rosso MN, Schiex T, Smant G, Weissenbach J, Wincker P. Genome sequence of the metazoan plant-parasitic nematode Meloidogyne incognita. Nature Biotechnology, 2008, 26(8): 909-915. DOI:10.1038/nbt.1482 |
| [22] | Zasada IA, Halbrendt JM, Kokalis-Burelle N, LaMondia J, McKenry MV, Noling JW. Managing nematodes without methyl bromide. Annual Review of Phytopathology, 2010, 48: 311-328. DOI:10.1146/annurev-phyto-073009-114425 |
| [23] | Kim M, Koh HS, Ichikawa T, Fukami H, Ishii S. Antifeedant of barnyard grass against the brown planthopper, Nilaparvata lugens (STÅL) (Homoptera:Delphacidae). Applied Entomology and Zoology, 1975, 10(2): 116-122. DOI:10.1303/aez.10.116 |
| [24] | Katsuhara M, Sakano K, Sato M, Kawakita H, Kawabe S. Distribution and production of trans-aconitic acid in barnyard grass (Echinochloa crus-galli var. oryzicola) as putative antifeedant against brown planthoppers. Plant and Cell Physiology, 1993, 34(2): 251-254. |
| [25] | Cheng XY, Zhu LL, He GC. Towards understanding of molecular interactions between rice and the brown planthopper. Molecular Plant, 2013, 6(3): 621-634. DOI:10.1093/mp/sst030 |
| [26] | Kim CS, Alamgir KM, Matsumoto S, Tebayashi SI, Koh HS. Antifeedants of Indian barnyard millet, Echinochloa frumentacea link, against brown planthopper, Nilaparvata lugens (Stål). Zeitschrift für Naturforschung C, 2008, 63(9/10): 755-760. |
| [27] | Misra S, Sanyal T, Sarkar D, Bhattacharya PK, Ghosh DK. Evaluation of antileishmanial activity of trans-aconitic acid. Biochemical Medicine and Metabolic Biology, 1989, 42(3): 171-178. DOI:10.1016/0885-4505(89)90052-2 |
| [28] | Kar S, Kar K, Bhattacharya PK, Ghosh DK. Experimental visceral leishmaniasis:role of trans-aconitic acid in combined chemotherapy. Antimicrobial Agents and Chemotherapy, 1993, 37(11): 2459-2465. DOI:10.1128/AAC.37.11.2459 |
| [29] | de Faria Garcia E, de Oliveira MA, Godin AM, Ferreira WC, Bastos LFS, de Matos Coelho M, Braga FC. Antiedematogenic activity and phytochemical composition of preparations from Echinodorus grandiflorus leaves. Phytomedicine, 2010, 18(1): 80-86. DOI:10.1016/j.phymed.2010.05.008 |
| [30] | de Faria Garcia E, de Oliveira MA, Dourado LPA, de Souza DG, Teixeira MM, Braga FC. In Vitro TNF-α inhibition elicited by extracts from Echinodorus grandiflorus leaves and correlation with their phytochemical composition. Planta Medica, 2016, 82(4): 337-343. |
| [31] | Gutierrez EN, Lamberti V. Preparation of aconitic acid. USA: 4123459. 1978-10-31. |
| [32] | He J, Wang JP, Yin W, Shao XH, Zheng HJ, Li MS, Zhao YW, Sun M, Wang SY, Yu ZN. Complete genome sequence of Bacillus thuringiensis subsp. chinensis strain CT-43. Journal of Bacteriology, 2011, 193(13): 3407-3408. DOI:10.1128/JB.05085-11 |
| [33] | Zheng C, Wang JP, Luo YC, Fu Y, Su JM, He J. Highly efficient enzymatic preparation of c-di-AMP using the diadenylate cyclase DisA from Bacillus thuringiensis. Enzyme and Microbial Technology, 2013, 52(6/7): 319-324. |
| [34] | Saffran M, Prado JL. Inhibition of aconitase by trans-aconitate. The Journal of Biological Chemistry, 1949, 180(3): 1301-1309. DOI:10.1016/S0021-9258(19)51244-3 |
| [35] | Cai H, Clarke S. A novel methyltransferase catalyzes the methyl esterification of trans-aconitate in Escherichia coli. The Journal of Biological Chemistry, 1999, 274(19): 13470-13479. DOI:10.1074/jbc.274.19.13470 |
 2021, Vol. 61
2021, Vol. 61




