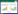中国科学院微生物研究所,中国微生物学会,中国菌物学会
文章信息
- 马建荣, 余永红, 陈艺彩, 鄢明峰, 张文彬. 2021
- Jianrong Ma, Yonghong Yu, Yicai Chen, Mingfeng Yan, Wenbin Zhang. 2021
- 细菌中酶蛋白硫辛酰化途径研究进展
- Advances in protein lipoylation pathways in bacteria
- 微生物学报, 61(8): 2278-2293
- Acta Microbiologica Sinica, 61(8): 2278-2293
-
文章历史
- 收稿日期:2020-09-29
- 修回日期:2021-01-16
- 网络出版日期:2021-07-29
2. 华南农业大学生命科学学院, 广东省农业生物蛋白质功能与调控重点实验室, 广东 广州 510642
2. Guangdong Provincial Key Laboratory of Protein Function and Regulation in Agricultural Organisms, College of Life Sciences, South China Agricultural University, Guangzhou 510642, Guangdong Province, China
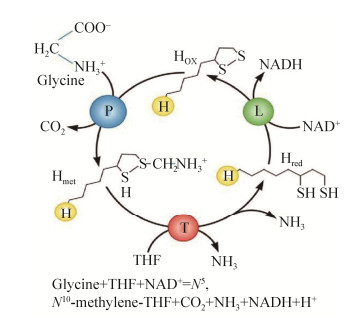
硫辛酸(lipoic acid)是在6位和8位碳原子之间连接两个硫原子的八碳饱和脂肪酸,其化学名称为1, 2-二硫代戊环-3-戊酸(图 1)[1]。硫辛酸具有氧化型(LA)和还原型(DHLA)之分,既具水溶性又具脂溶性,还具有强抗氧化性,能直接清除活性氧、螯合重金属离子,还能再生其他抗氧化剂如维生素C、谷胱甘肽、硫氧还原蛋白等,被誉为“万能抗氧化剂”[2]。由于硫辛酸具有缓解肌体过度疲劳、延缓衰老、预防记忆力衰退等功效,在美容养颜、减肥等方面也有功效,同时硫辛酸也应用于糖尿病、脑和神经退化性疾病以及肝脏类疾病治疗等,因此硫辛酸在保健食品、化妆品和药品等领域都具有广泛的应用前景[3-5]。硫辛酸属于维生素B类化合物,在生物体内作为辅因子广泛存在。硫辛酸与酶蛋白中赖氨酸的ε-氨基共价结合,以酰胺键形式连接,完成酶蛋白翻译后硫辛酰化修饰过程,并维持酶蛋白的催化活性。模式生物大肠杆菌(Escherichia coli)中酶蛋白硫辛酰化途径研究最早,发现了硫辛酸从头合成化途径和硫辛酸补救合成途径。但随着研究深入,发现不同细菌硫辛酰化途径具有较高的多样性,革兰氏阳性细菌、革兰氏阴性细菌和古生菌中酶蛋白硫辛酰化途径差异明显,两种途径中催化酶类、载体蛋白都具有多样性,而部分细菌中则只有一种硫辛酰化途径。由于硫辛酸是细菌生长的必需辅因子,研究细菌中硫辛酰化机制不仅可发现新的抗菌药物靶点,还可为生物法高产硫辛酸提供理论基础[6]。为此,本文将对细菌中酶蛋白硫辛酰化途径的研究进展进行综述。
|
| 图 1 辛酸、硫辛酸和二氢硫辛酸的结构 Figure 1 The chemical structures of octanoic acid, lipoic acid and dihydrolipoic acid. |
1 硫辛酸依赖的多酶复合体
虽然硫辛酸作为一种抗氧化剂被广泛使用,但研究最清楚的是硫辛酸作为辅基参与α-酮酸脱氢酶复合体(α-oxo acid dehydrogenase complex)的催化反应,包括丙酮酸脱氢酶(pyruvate dehydrogenase,PDH)、α-酮戊二酸脱氢酶(α-oxoglutarate dehydrogenase,OGDH)、支链酮酸脱氢酶(branched-chain ketoacid dehydrogenase,BKDH)和3-羟基丁酮脱氢酶(acetoin dehydrogenase,ADH)等4种复合酶体[7-8]。其中丙酮酸脱氢酶(PDH)和α-酮戊二酸脱氢酶(OGDH)参与生物体的能量代谢,还可以合成中间代谢产物乙酰-CoA、琥珀酰-CoA等,作为前体用于氨基酸、脂肪酸的合成。支链酮酸脱氢酶(BCDH)可催化支链酮酸脱羧生成支链脂酰-CoA,作为前体用于支链脂肪酸合成[7-8]。部分细菌还编码3-羟基丁酮脱氢酶(ADH),催化3-羟基丁酮分解为乙酰-CoA和乙醛,提供生长所需的能量和碳源[9]。
α-酮酸脱氢酶复合体都含有多拷贝的三种亚基E1、E2和E3,每种亚基都催化部分反应,而硫辛酸的羧基与E2亚基上保守结构域的赖氨酸残基中ε-氨基共价结合,催化反应中硫辛酸起在多酶复合体的各个活性中心之间传递中间产物的作用[9]。α-酮酸脱氢酶复合体催化反应包括:E1亚基催化焦磷酸硫胺素依赖的α-酮酸氧化脱羧,同时还原性酰基化E2上的硫辛酸基团;E2亚基催化酰基从硫辛酸基团上转移给CoA,产生酰基CoA;E3亚基催化硫辛酸基团再氧化,形成氧化态硫辛酸(图 2)[10]。在α-酮酸脱氢酶复合体中,E2亚基处于复合体中心,E1和E3通过非共价键与之紧密结合[11]。E1和E2具有底物专一性,E3在同种细菌中为多种α-酮酸脱氢酶复合体所共有[12]。
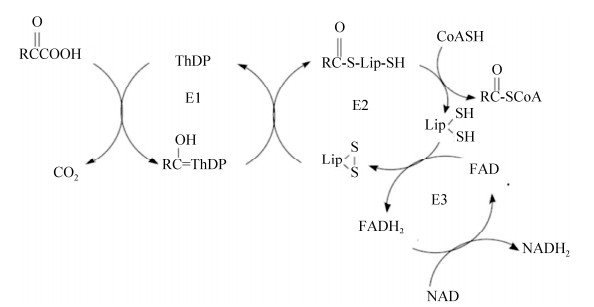
另外,硫辛酸作为辅因子也在甘氨酸裂解系统(glycine cleavage system,GCS)中发挥重要作用。甘氨酸裂解系统催化甘氨酸裂解为CO2和NH3,同时催化合成一碳单位N5, N10-亚甲基四氢叶酸(THF)。甘氨酸裂解系统由四种蛋白(GcvP、GcvH、GcvT和GcvL)组成,其中GcvH是硫辛酸的载体蛋白,GcvP催化甘氨酸脱羧,并将剩余的氨基甲基连接到硫辛酸的巯基上,GcvT分解氨基甲基,将一碳单位转移到四氢叶酸(THF)上并释放氨,而GcvL负责GcvH上的二氢硫辛酸再氧化生成硫辛酸,与α-酮酸脱氢酶复合体的E3亚基类似(图 3)[13]。
2 硫辛酸结构域与GcvH蛋白
在α-酮酸脱氢酶复合体中E2亚基含多个结构域,从N端开始依次是3个硫辛酸结构域(lipoyl domain,LD)、亚基结合结构域和酰基转移酶结构域,各结构域之间由富含丙氨酸、脯氨酸和带电氨基酸组成的多肽链(25–30氨基酸残基)隔开(图 4-A)[12, 15]。硫辛酸结构域约含有80个氨基酸残基,不同来源的硫辛酸结构域蛋白具有保守的空间结构(PDB code 1QJQ),包含2个由4条β-折叠链组成的β-片层,其序列N端和C端残基靠近,而与硫辛酸结合的赖氨酸残基处在β-转角中(图 4-B)[16]。硫辛酸结构域直接参与催化反应中底物传递和活性偶联,也是E2亚基在翻译后被硫辛酰化修饰的识别位点。另外,体内体外的研究还证明硫辛酸结构域与E2亚基一样,可作为底物被不同酶催化发生硫辛酰基化修饰[17-18]。

|
| 图 4 大肠杆菌丙酮酸脱氢酶E2亚基(A)、硫辛酸结构域(B)和GcvH蛋白(C) Figure 4 The E2 subunit of pyruvate dehydrogenase (A), lipoyl domain (B) and GcvH protein in E. coli. The PDB codes of E. coli lipoyl domain and GcvH are 1QJQ and 3AB9, respectively. |
大肠杆菌甘氨酸裂解系统中GcvH (EcGcvH)蛋白也需要硫辛酰化修饰后才能发挥作用。EcGcvH (PDB code 3AB9)具有129个氨基酸,其序列与丙酮酸脱氢酶E2亚基的硫辛酸结构域序列相似性较低(氨基酸序列一致性 < 20%),但具有高度相似的空间结构,也含有保守的赖氨酸残基与硫辛酸共价结合(图 4-C)[7, 19]。大肠杆菌中硫辛酰化修饰可直接在GcvH蛋白或α-酮酸脱氢酶复合物E2亚基的硫辛酸结构域上完成,但枯草芽孢杆菌只能首先在GcvH上完成硫辛酰化修饰,再转移到E2亚基的硫辛酸结构域上(见下文),因此其GcvH不仅参与甘氨酸裂解,还负责完成硫辛酸转运[20]。具有类似双功能(甘氨酸裂解和硫辛酰化修饰)的GcvH被陆续报道,如Aquifex aeolicus中5个GcvH同源蛋白中,2个(GcvH1和GcvH4)具有双功能,而其他3个只在甘氨酸裂解中发挥作用[21]。
3 大肠杆菌中酶蛋白硫辛酰化途径模式生物大肠杆菌有两种酶蛋白硫辛酰化途径:硫辛酸从头合成途径和硫辛酸补救合成途径,两种途径相互补充(图 5)[13]。硫辛酸从头合成途径是指大肠杆菌以辛酰ACP (acyl carrier protein)为底物从头合成硫辛酸,并完成酶蛋白硫辛酰化修饰;硫辛酸补救合成途径是指细菌从生长环境中直接获得硫辛酸,并完成酶蛋白硫辛酰化修饰。当生长环境中有硫辛酸时,细菌首先利用外源硫辛酸完成酶蛋白修饰;而当环境中缺乏硫辛酸时,细菌则通过从头合成途径合成硫辛酸供给细胞利用。
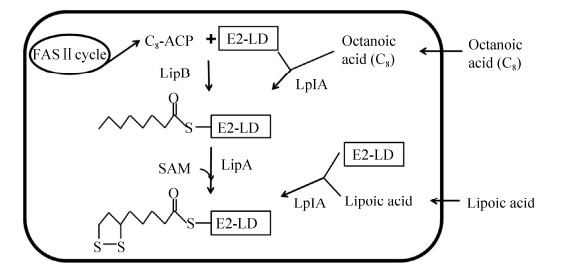
|
| 图 5 大肠杆菌中酶蛋白硫辛酰化途径 Figure 5 Protein lipoylation pathways in E. coli. E2-LD refers to the lipoyl domain in the E2 subunit of α-oxo acid dehydrogenase. |
3.1 硫辛酸从头合成途径
大肠杆菌中硫辛酸从头合成途径以脂肪酸合成代谢中间产物辛酰ACP为底物,在辛酰转移酶(LipB)催化下,将辛酰基团转移到α-酮酸脱氢酶E2亚基的硫辛酸结构域上,进一步在硫辛酸合成酶(LipA)催化下,在辛酰基团的6位和8位碳原子之间连接两个硫原子形成硫辛酰化蛋白亚基(图 5)[13, 22-23]。
大肠杆菌LipB是一个大约为29 kDa的单体蛋白(PDB code 2QHV),能够以辛酰ACP为底物,催化辛酰基团转移并与LipB中第169位半胱氨酸(Cys169)生成硫脂键,而后进一步将辛酰基团转到E2亚基(或GcvH蛋白)中,与硫辛酸结构域中赖氨酸相连形成酰胺键[22, 24]。而晶体结构研究也证明LipB为新型半胱氨酸/赖氨酸二元酰基转移酶,半胱氨酸/赖氨酸残基在催化过程中发挥酸/碱催化剂作用[17, 25]。
大肠杆菌LipA属于S-腺苷甲硫氨酸(SAM)自由基酶超级家族,含有由3个保守的半胱氨酸组成的CX3CX2C簇,而N端保守的CX4CX5C辅助簇在硫辛酸合成中也发挥重要作用(PDB code 5EXJ)[26]。大肠杆菌LipA以同质二聚体形式存在,含有SAM自由基[4Fe-4S]簇和辅助[4Fe-4S]簇,而1个LipA单体的辅助[4Fe-4S]簇为硫辛酸合成提供2个硫原子[27]。LipA催化硫辛酸合成可分为两步进行,首先[4Fe-4S]簇裂解S-腺苷甲硫氨酸生成甲硫氨酸和5ʹ-脱氧腺苷自由基,后者攻击辛酰基团上第6位碳原子,由此产生的碳中心自由基攻击LipA的辅助簇,与辛酰基团生成共价连接;而第2个5ʹ-脱氧腺苷自由基则攻击辛酰基团的第8位碳原子,生成的碳中心自由基攻击第2个辅助簇,并最终生成还原型的二氢硫辛酸[28]。
大肠杆菌LipA催化硫辛酸合成后,其辅助[4Fe-4S]簇可由Fe-S簇载体蛋白NfuA介导再生[29]。大肠杆菌NfuA(EcNfuA)蛋白的C端半胱氨酸残基在Fe-S簇生成中发挥作用,而N端A-type域在与LipA互作中发挥关键作用[30-31]。结核分枝杆菌(Mycobacterium tuberculosis)来源的MtNfuA也能催化大肠杆菌LipA的辅助簇再生,但金黄色葡萄球菌(Staphylococcus aureus)的Nfu (SaNfu)由于缺乏A-type域不能促进再生,而将EcNfuA的N端结构域连接到SaNfu上,则能恢复SaNfu活性[32]。
3.2 硫辛酸补救合成途径大肠杆菌中硫辛酸补救合成途径依赖于硫辛酸连接酶LplA。该反应分两步催化,首先LplA催化硫辛酸与ATP结合生成活化的中间产物硫辛酰-AMP,并释放出焦磷酸;而后LplA催化硫辛酰基团从硫辛酰-AMP转移到α-酮酸脱氢酶复合体E2亚基的硫辛酸结构域(或GcvH)上,与保守的赖氨酸残基中ε-氨基生成酰胺键,并伴随AMP的释放[24, 33](图 5)。
大肠杆菌lplA是第一个被发现的硫辛酸连接酶基因,其编码产物也是第一个被纯化出来的硫辛酸连接酶。大肠杆菌LplA为单体蛋白,分子量为38 kDa,可独立催化硫辛酸的活化和转移两个反应,但其在体内和体外对辛酸也有催化活性,具有底物广泛性[34]。虽然大肠杆菌LipB和硫辛酸连接酶LplA没有序列相似性,催化不同的化学反应,但两者具有相似的3D结构,大肠杆菌LipB对硫辛酰ACP也具有较弱的催化活性,而大肠杆菌LplA也有微弱的LipB功能[35]。
晶体结构分析发现大肠杆菌LplA (PDB code 3A7R)包含一个大的N末端结构域和一个小的C末端结构域,底物结合口袋存在于两结构域之间,硫辛酸通过疏水作用和羧基端的氢键作用与LplA结合[34]。比较不同来源的硫辛酸连接酶,发现LplA含有3个保守的基序(motifs),基序Ⅰ为(RRXXGGG XV(F/Y)HD),其中第1个Arg (R)在与硫辛酸结构域的相互作用过程中起重要作用;基序Ⅱ为(KhXGXA),其中Lys (K)是严格保守的氨基酸,在硫辛酰-AMP生成和硫辛酸转移过程中都发挥关键性作用;基序Ⅲ为(HXX(L/M)LXXX(D/N) LXXLXXhL),其中X代表任何氨基酸,而h代表疏水氨基酸(图 6)[33, 36]。这些保守区域的结构分析对酶活性鉴定具有重要的参考价值。
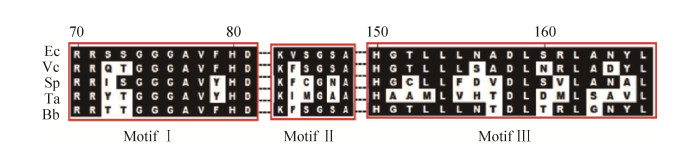
|
| 图 6 不同细菌来源的LplA序列比对 Figure 6 Sequence alignment of LplAs from different bacteria. Ec: E. coli (SWISS-PROT accession code: P32099); Ta: T. acidophilum (Q9HKT1); Vc: Vibrio cholerae (Q9KS71); Bb: Bdellovibrio bacteriovorus (P60809); Sp: S. pneumoniae (Q8DPR1). The residue numbering on the top refers to EcLplA. |
4 不同细菌中硫辛酸从头合成途径多样性
大肠杆菌中硫辛酰化途径关键酶同源蛋白在真菌、植物、原生生物和哺乳动物中不断被发现,揭示该途径在不同物种间广泛存在,尽管具体的催化机制可能有差别。但随着研究的深入,发现不同细菌中硫辛酸从头合成途径具有多样性[8]。革兰氏阳性细菌枯草芽孢杆菌(Bacillus subtilis)编码具有催化功能的大肠杆菌LipA同源蛋白,但基因组中没有大肠杆菌lipB同源基因[37]。生物信息学分析显示,枯草芽孢杆菌含有3个硫辛酸连接酶同源蛋白编码基因,其中LipM (BsLipM)为辛酰转移酶。虽然BsLipM和大肠杆菌LipB只有20%的序列一致性,但两者的催化功能和催化机制相似,以脂肪酸合成代谢中间产物辛酰ACP为底物,BsLipM将辛酰基团转移到其C150位点上,生成硫脂键相连的中间产物,而后进一步将辛酰基团转移到枯草芽孢杆菌GcvH中(图 7-A)[38]。BsLipM也能以辛酰-CoA为底物,催化辛酰基团转移到GcvH上,但活性较弱。BsLipM为新型辛酰转移酶,其中C150和K165是催化活性的关键氨基酸,而BsLipM同源蛋白广泛分布于厚壁菌门和蓝藻中[39]。但BsLipM不能将辛酰基团转移到α-酮酸脱氢酶复合物的E2亚基上,也说明枯草芽孢杆菌中还有其他酶参与硫辛酸的从头合成。
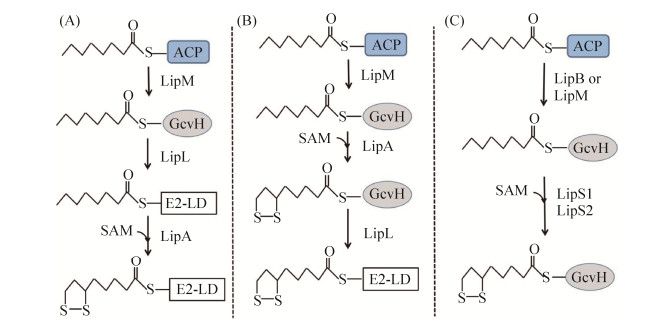
|
| 图 7 不同细菌中硫辛酸从头合成途径 Figure 7 The lipoic acid de novo synthetic pathways in different bacteria. A: B. subtilis; B: S. aureus; C: Thermococcus kodakarensis. E2-LD refers to the lipoyl domain in the E2 subunit of α-keto acid dehydrogenase. |
枯草芽孢杆菌中硫辛酸从头合成还需要LipL(BsLipL)参与,BslipL突变株是硫辛酸营养缺陷型,但培养基中即使添加硫辛酸也只能部分恢复△lipL的生长,说明BsLipL还影响硫辛酸补救合成途径(详见下文)[20]。而△lipL菌株无细胞提取物中硫辛酰化修饰的GcvH出现累积,但由于E2亚基不能被硫辛酸修饰,细胞内α-酮酸脱氢酶复合物没有活性。BslipL基因不能互补大肠杆菌lipB突变株,但大肠杆菌lipB能恢复△lipL菌株在基础培养基上生长,证明BsLipL和BsLipM在E2亚基的辛酰修饰中发挥作用[20]。在大肠杆菌中表达BsLipL,检测发现其C39和C150位发生硫辛酰或辛酰修饰。因此BsLipL是一种新的氨酰转移酶,以辛酰-GcvH为底物,催化辛酰基团转移到丙酮酸脱氢酶的E2亚基上,其中C39位点突变不影响催化活性,而C150为活性关键位点,与BsLipB和BsLipM类似,BsLipL具有Cys-Lys二元催化活性中心,但BsLipL与其他氨酰转移酶没有序列或结构的相似性[20]。而在枯草芽孢杆菌中,GcvH蛋白也是硫辛酸合成的关键蛋白,gcvH突变株为硫辛酸营养缺陷型(图 7-A)。
人类条件致病菌金黄色葡萄球菌(Staphylococcus aureus)也具有与枯草芽孢杆菌类似的硫辛酰化途径,其基因组编码硫辛酸合成关键酶SaLipM、SaLipA和SaLipL,其中SaLipM与BsLipM具有62%的序列一致性,也具有类似的功能,即催化辛酰基团从辛酰ACP转移到GcvH上[40]。但与枯草芽孢杆菌的硫辛酸从头合成步骤不同,金黄色葡萄球菌中硫辛酸合成酶SaLipA以辛酰-GcvH为底物,催化在C6和C8位连接2个硫原子生产硫辛酰-GcvH,而后氨酰转移酶SaLipL再将硫辛酰基团转移不同脱氢酶复合体的E2亚基上,完成内源性硫辛酰化修饰过程(图 7-B)[41]。金黄色葡萄球菌中硫辛酸从头合成途径在对寄主的全身性感染中发挥关键作用[42]。
嗜热古生菌Thermococcus kodakarensis生长不需要添加硫辛酸,但其基因组不编码LipA同源蛋白,推测该菌具有新的硫辛酸从头合成途径。生物信息学分析发现2个标注为生物素合成酶TK2109 (LipS1)和TK2248 (LipS2)可能参与硫辛酸合成代谢,但LipS1和LipS2与大肠杆菌LipA、嗜热古生菌Sulfolobus solfataricus LipA (SSO3158)的序列一致性都低于20%。lipS1敲除突变株和lipS2敲除突变株均为硫辛酸的营养缺陷型,而体外实验发现单独LipS1或LipS2都不能以催化辛酰-GcvH生成硫辛酰产物,但LipS1和LipS2能共同催化反应发生,生成硫辛酰-GcvH,证明两者都是催化活性必需的(图 7-C),而LipS1和LipS2同源蛋白在嗜热古生菌中广泛存在[43]。但Thermococcus kodakarensis中辛酰转移酶LipB或者LipM同源蛋白还没有被鉴定,而LipS1和LipS2催化生成硫辛酰-GcvH后,硫辛酰基团如何转移到α-酮酸脱氢酶复合体也还不清楚。
5 不同细菌中硫辛酸补救合成途径的多样性许多细菌可以从环境中“觅食(scavenge)”硫辛酸,但培养基或肠道环境中硫辛酸通常与载体蛋白相结合,而部分细菌可编码硫辛酸酰胺酶(lipoamidase)将硫辛酸解离下来,并以自由扩散方式进入细胞内,再通过硫辛酸连接酶催化生成硫辛酰-GcvH或硫辛酰-E2亚基,完成酶蛋白的硫辛酰化修饰[44-45]。虽然与模式生物大肠杆菌类似的硫辛酸补救合成途径被陆续报道,但不同细菌中该途径也有较大差异。
生物信息学分析显示,枯草芽孢杆菌基因组编码3个硫辛酰蛋白连接酶同源蛋白,但只有BsLplJ具有硫辛酸连接酶活性。BsLplJ与大肠杆菌LplA (EcLplA)序列一致性为33%,体外检测发现对硫辛酸和辛酸都具有催化活性,可以将其连接到GcvH上。α-酮戊二酸脱氢酶(OGDH)复合体中E2亚基也是BsLplJ的催化底物,BsLplJ可利用外源硫辛酸完成OGDH的硫辛酰化修饰(图 8-A)[20, 39]。但枯草芽孢杆菌BslipL突变株在添加硫辛酸的培养基上也不能完全恢复生长,说明BsLipL还参与硫辛酸补救合成过程。BslipL突变株中硫辛酰化GcvH出现累积,表明BsLipL在催化硫辛酰基团转移到E2亚基中发挥关键作用,同时BsLipL还能将OGDH上的硫辛酰基团转移到丙酮酸脱氢酶(PDH)、支链酮酸脱氢酶(BKDH)的E2亚基上,完成硫辛酰化修饰过程(图 8-A)[46]。与大肠杆菌LplA不同,BsLplJ只能催化GcvH和OGDH的E2亚基,而不能直接完成PDH和BKDH的E2亚基硫辛酰化修饰,将不同多酶复合体的E2亚基、GcvH序列比对发现,PDH和BKDH的E2亚基中硫辛酰化修饰位点Lys (K)上游第3个氨基酸由保守的谷氨酸(酸性氨基酸)分别变成了谷氨酰胺、甲硫氨酸,将OGDH中该位点的谷氨酸突变为谷氨酰胺则失去活性,而将PDH中谷氨酰胺突变为谷氨酸则恢复活性,充分证明该位点是BsLplJ底物识别的关键位点[46]。
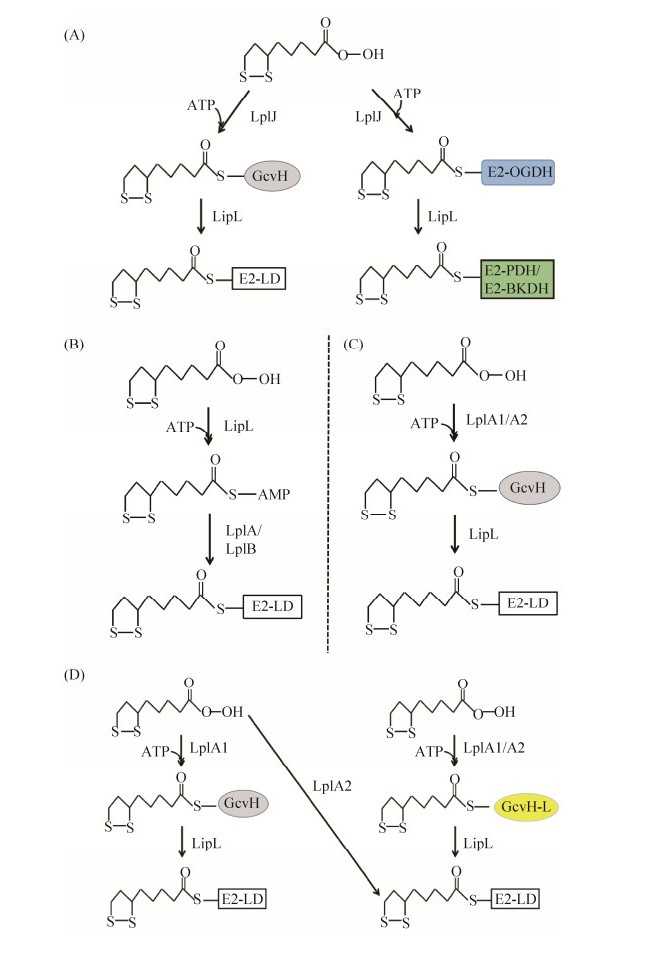
|
| 图 8 不同细菌中硫辛酸补救合成途径 Figure 8 The lipoic acid salvage pathways in different bacteria. A: B. subtilis; B: T. acidophilum; C: L. monocytogenes; D: S. aureus. E2-LD refers to the lipoyl domain in the E2 subunit of α-oxoacid dehydrogenase; OGDH: α-oxoglutarate dehydrogenase; PDH: pyruvate dehydrogenase; BKDH: branched-chain ketoacid dehydrogenas. |
古生菌Thermoplasma acidophilum基因组编码2个大肠杆菌LplA同源蛋白TaLplA和TaLplB,但只有同时表达TalplA和TalplB才能恢复大肠杆菌lplA和lipA双突变株的生长[47]。与EcLplA相比,TaLplA缺乏C端结构域(C-terminal domain,CTD),但可催化硫辛酸与ATP连接生成硫辛酰-AMP,并稳定结合在TaLplA蛋白的U型口袋中(PDB code2ARS)[33]。但TaLplA不能进一步将硫辛酸从硫辛酰-AMP连接到载体蛋白上,证明EcLplA中C端结构域在催化硫辛酸第二步转移中发挥关键作用(图 8-B)。而只有当TaLplB与TaLplA协作,才能进一步将硫辛酰基团转移到载体蛋白上,但TaLplB不与硫辛酸结构域直接相互作用。以上结果说明硫辛酸连接酶可以拆分为2个部分,当两者同时存在时,即可完成催化作用(图 8-B)[47-48]。同时生物信息学分析发现古生菌中硫辛酸连接酶都是异二聚体[47]。
天蓝色链霉菌(Streptomyces coelicolor)基因组只编码一个硫辛酸连接酶ScLplA,但与已知的硫辛酸连接酶序列一致性较低(< 25%),而且硫辛酸连接酶保守的C端结构域出现在ScLplA的N端[49]。体内异体遗传互补和体外活性分析都证明ScLplA具有硫辛酸连接酶活性。将2个结构域模拟EcLplA序列重排后的蛋白依然具有硫辛酸连接酶活性,两个结构域在催化硫辛酰-AMP形成过程中都发挥作用,而仅大结构域可催化将硫辛酸从硫辛酰-AMP连接到受体蛋白上[49]。
单增李斯特菌(Listeria monocytogenes)是革兰氏阳性厚壁菌门的人类致病菌,为硫辛酸营养缺陷型,需要吸收外源硫辛酸才能生长[50]。单增李斯特菌编码两个硫辛酸连接酶LmLplA1和LmLplA2,在基础培养基中添加25 μmol/L硫辛酸时,表达LmlplA1或LmlplA2都能恢复大肠杆菌lplA和lipA双突变株生长,但在基础培养基中只添加极微量硫辛酸(25 nmol/L)时,只有表达LmlplA1能恢复大肠杆菌lplA和lipA双突变株的生长,表明LmLplA1的催化活性要显著高于LmLplA2[44]。体外实验证明LmLplA1与枯草芽孢杆菌BsLipJ类似,能催化硫辛酸连接到GcvH上,但不能将脱氢酶复合物E2亚基硫辛酰化(图 8-C)[44]。单增李斯特菌还编码氨酰转移酶LipL (LmLipL),LmLipL能与丙酮酸脱氢酶复合物结合,可逆性催化硫辛酰基团从GcvH蛋白转移到脱氢酶复合物的E2亚基上,说明LmLipL在硫辛酸的吸收利用过程中也是必不可少的(图 8-C)。单增李斯特菌也编码以Mn2+离子为辅因子的新型硫辛酸酰胺酶Lpd,将连接在蛋白上的硫辛酰基团解离,并被LmLplA1催化吸收[44]。
与大肠杆菌类似,金黄色葡萄球菌也同时具有硫辛酸从头合成途径和硫辛酸补救合成途径,其中从头合成途径是感染心脏时所必需的,而补救合成途径在感染肾脏过程中是关键的[40, 42]。与单增李斯特菌类似,金黄色葡萄球菌也编码2个硫辛酸连接酶SaLplA1和SaLplA2,两者都具有催化活性[40, 51]。金黄色葡萄球菌还编码2种H蛋白,甘氨酸裂解系统的SaGcvH和同源蛋白SaGcvH-L,后者编码基因与SalplA2位于同一个操纵子中。SaLplA1能将硫辛酸连接到两种H蛋白(SaGcvH和SaGcvH-L)上,而SaLplA2能将硫辛酸连接到酮酸脱氢酶复合体的E2亚基上,也能将硫辛酸连接到SaGcvH-L蛋白上。SaGcvH和SaGcvH-L上的硫辛酰基团可以进一步被硫辛酸转移酶SaLipL催化连接到E2亚基上(图 8-C)[41]。SaLplA1和SaLplA2都能以辛酸作为底物,催化连接到SaGcvH和SaGcvH-L上,而后在SaLipA催化下生产硫辛酸,因此金黄色葡萄球菌在感染寄主过程中有多种方式完成硫辛酰化修饰以维持生长[52]。
猪肺炎支原体(Mycoplasma hyopneumoniae)中Lpl蛋白具有硫辛酸连接酶活性,体外可将硫辛酸活化为硫辛酰-AMP,并进一步转移到其GcvH蛋白上。酶活性分析发现其K56残基与硫辛酸共价连接,其N端结构域含有完整的催化活性[53]。
6 总结和展望虽然半个多世纪前我们已经了解硫辛酸的结构及其在细菌代谢中的作用,但细菌中酶蛋白硫辛酰化途径在近10多年才被逐渐研究清楚[7]。模式生物大肠杆菌中酶蛋白硫辛酰化途径研究较为清楚,包括依赖于LipB-LipA的硫辛酸从头合成途径和依赖于LplA的硫辛酸补救合成途径,相关催化酶作用机制也比较透彻,为研究不同细菌中酶蛋白硫辛酰化途径奠定了重要的基础。但不同细菌中硫辛酰化途径具有较高的多样性(表 1),革兰氏阳性细菌中LipL和GcvH蛋白在2种硫辛酰化途径中都发挥重要作用,而硫辛酸补救合成途径中可能有多个LplA同源蛋白参与。古生菌中硫辛酰化途径研究报道较少,其从头合成途径中关键的硫辛酸合成酶LipS1/LipS2在嗜热古生菌的基因组中较为广泛存在,而许多嗜盐古生菌基因组编码LipA同源蛋白,而古生菌硫辛酸补救合成途径中硫辛酸连接酶通常含有LplA和LplB两部分(异二聚体结构)。
| Different bacteria | Lipoic acid de novo synthetic pathway | Lipoic acid salvage pathway |
| G– | ||
| Escherichia coli | LipB-LipA pathway Using octanoyl-ACP as a substrate, LipB catalyzes the formation of octanoyl-LD, then LipA catalyzes the formation of lipoyl-LD |
LplA LplA catalyzes the formation of lipoyl-LD with free lipioc acid |
| Xanthomonas campestris | No | |
| G+ | ||
| Bacillus subtilis | LipM-LipL-LipA pathway Using octanoyl-ACP as a substrate, LipM catalyzes the formation of octanoyl-GcvH, then LipL transfers the octanoyl group to LD, lastly LipA catalyzes the formation of lipoyl-LD |
LplJ-LipL pathway (1) LplJ catalyzes the formation of lipoyl-GcvH with free lipioc acid, then LipL transfers the lipoyl group to LD (2) LplJ catalyzes the formation of lipoyl-LD of OGDH with free lipioc acid, then LipL transfers the lipoyl group to LD of PDH and BKDH |
| Staphylococcus aureus | LipM-LipA-LipL pathway Using octanoyl-ACP as a substrate, LipM catalyzes the formation of octanoyl-GcvH, then LipA catalyzes the formation of lipoyl-GcvH, lastly LipL transfers lipoyl group to LD |
LplA1/LplA2-LipL pathway (1) LplA1 catalyzes the formation of lipoyl- GcvH (or -GcvH-L) with free lipioc acid, then LipL transfers the lipoyl group to LD (2) LplA2 catalyzes the formation of lipoyl- GcvH-L (or-LD), then LipL transfers the lipoyl group to LD |
| Listeria monocytogenes | No | LplA1/LplA2-LipL pathway LplA1 or LplA2 catalyzes the formation of lipoyl-GcvH with free lipioc acid, then LipL transfers the lipoyl group to LD |
| Streptomyces coelicolor | Unknown | ScLplA |
| Archaea | ||
| Thermoplasma acidophilum | Unknown | Ta LplA-Ta LplB LplA and LplB catalyze the formation of lipoyl-LD with free lipioc acid |
| Thermococcus kodakarensis | LipB(LipM)-LipS1/LipS2 pathway Using octanoyl-ACP as a substrate, LipB (or LipM) catalyzes the formation of octanoyl-GcvH, then LipS1/LipS2 catalyze the formation of lipoyl-GcvH |
Unknown |
| Others | ||
| Mycoplasma hyopneumoniae | Unknown | Lpl Lpl catalyzes the formation of lipoyl-GcvH with free lipioc acid |
| Mycobacterium tuberculosis | LipB-LipA pathway Using octanoyl-ACP as a substrate, LipB catalyzes the formation of octanoyl-LD, then LipA catalyzes the formation of lipoyl-LD |
No |
| G+: Gram positive bacteria; G–: Gram negative bacteria; LD: lipoyl domain of E2 subunit in α-oxo acid dehydrogenase; GcvH: the H protein in Glycine cleavage system; ACP: acyl carrier protein. | ||
硫辛酸作为细胞内重要的辅因子,对细菌生长、致病性等方面具有广泛影响,因此一些细菌不仅可以从头合成硫辛酸,完成内源性硫辛酰化修饰,还可编码硫辛酸酰胺酶获取环境中的非自由硫辛酸,利用补救合成途径完成硫辛酰化修饰过程。但部分细菌只含有一种硫辛酰化途径。结核分枝杆菌(Mycobacterium tuberculosis)中没有硫辛酸补救合成途径,只能通过LipB-LipA途径从头合成硫辛酸,因此LipB和LipA被认为是开发抗结核药物的理想靶标[6, 54]。植物病原菌野油菜黄单胞菌(Xanthomonas campestris)基因组编码同源性XcLipB-XcLipA,并催化硫辛酸从头合成,但野油菜黄单胞菌中没有大肠杆菌硫辛酸连接酶LplA同源蛋白,因此XclipA和XclipB都是必需基因,只有在野油菜黄单胞菌中表达大肠杆菌lplA后才能获得XclipA或XclipB的敲除突变株,而XclipA敲除突变株对寄主植物的致病力几乎完全丧失,证明在只有硫辛酸从头合成途径的病原菌中,硫辛酸合成酶LipA可能是良好的抗菌药物筛选靶点[55]。与之类似,在仅含有硫辛酸补救合成途径的细菌(如单增李斯特菌)中,其硫辛酰化修饰过程的酶类是菌体生长所必需,也可作为抗菌药物筛选的靶点[50]。
由于硫辛酸在食品、化妆品和药物等方面都具有非常高的经济价值,但硫辛酸化学合成方法(己二酸法和环己酮法)都有步骤繁琐、工艺复杂且均采用化工原料污染环境等弊端,因此如何利用生物法高效生产硫辛酸是当前的研究热点,而对硫辛酸合成途径的研究可为获得高产硫辛酸的基因工程菌奠定基础[56]。Sun等根据大肠杆菌中硫辛酸从头合成途径,在E. coli BL21中利用不同策略过量表达硫辛酸结构域LipD、硫辛酸合成酶LipA和连接酶LplA,并添加辛酸作为硫辛酸合成前体,结果显示基因工程菌中硫辛酸产量提高了200倍[57]。利用不同来源的硫辛酸合成酶或连接酶催化活性差异,可进一步在不同细胞体系中构建硫辛酸高产菌株,为工业化应用提供可能性。
| [1] | Reed KE, Cronan JE. Lipoic acid metabolism in Escherichia coli: sequencing and functional characterization of the lipA and lipB genes. Journal of Bacteriology, 1993, 175(5): 1325-1336. DOI:10.1128/jb.175.5.1325-1336.1993 |
| [2] |
Liao DF, Chen JW, Xie P, Zhu ZQ, Xu R. Studies on the antioxidation effects of alpha-lipoic acid and dihydrolipoic acid. Journal of East China Normal University: Natural Science, 2007(2): 87-92, 136.
(in Chinese) 廖德丰, 陈季武, 谢萍, 朱振勤, 徐容. α-硫辛酸和二氢硫辛酸的抗氧化作用. 华东师范大学学报: 自然科学版, 2007(2): 87-92, 136. |
| [3] | Tajima K, Ikeda K, Chang HY, Chang CH, Yoneshiro T, Oguri Y, Jun H, Wu J, Ishihama Y, Kajimura S. Mitochondrial lipoylation integrates age-associated decline in brown fat thermogenesis. Nature Metabolism, 2019, 1(9): 886-898. DOI:10.1038/s42255-019-0106-z |
| [4] |
Liu Y, Wang EX, Pan CH, Dong XT, Ding MZ. Biosynthesis of a-lipoic acid in Gluconobacter oxydans increases the production of vitamin C by one-step fermentation. Chinese Journal of Biotechnology, 2019, 35(7): 1266-1276.
(in Chinese) 刘宇, 王恩旭, 潘才惠, 董秀涛, 丁明珠. 氧化葡萄糖酸杆菌中硫辛酸合成模块对维生素C一步混菌发酵的影响. 生物工程学报, 2019, 35(7): 1266-1276. |
| [5] | Hayakawa S, Kawamura M, Sato T, Hirano T, Kikuchi T, Watanabe A, Fujimura S. An α-Lipoic acid derivative, and anti-ROS agent, prevents the acquisition of multi-drug resistance in clinical isolates of Pseudomonas aeruginosa. Journal of Infection and Chemotherapy, 2019, 25(1): 28-33. DOI:10.1016/j.jiac.2018.10.003 |
| [6] | Lanz ND, Lee KH, Horstmann AK, Pandelia ME, Cicchillo RM, Krebs C, Booker SJ. Characterization of lipoyl synthase from Mycobacterium tuberculosis. Biochemistry, 2016, 55(9): 1372-1383. DOI:10.1021/acs.biochem.5b01216 |
| [7] | Cronan JE. Advances in synthesis of biotin and assembly of lipoic acid. Current Opinion in Chemical Biology, 2018, 47: 60-66. DOI:10.1016/j.cbpa.2018.08.004 |
| [8] | Cronan JE. Assembly of lipoic acid on its cognate enzymes: an extraordinary and essential biosynthetic pathway. Microbiology and Molecular Biology Reviews, 2016, 80(2): 429-450. DOI:10.1128/MMBR.00073-15 |
| [9] | Spalding MD, Prigge ST. Lipoic acid metabolism in microbial pathogens. Microbiology and Molecular Biology Reviews, 2010, 74(2): 200-228. |
| [10] | Cronan JE. Progress in the enzymology of the mitochondrial diseases of lipoic acid requiring enzymes. Frontiers in Genetics, 2020, 11: 510. DOI:10.3389/fgene.2020.00510 |
| [11] | Perham RN. Swinging arms and swinging domains in multifunctional enzymes: catalytic machines for multistep reactions. Annual Review of Biochemistry, 2000, 69: 961-1004. DOI:10.1146/annurev.biochem.69.1.961 |
| [12] | Perham RN. Domains, motifs, and linkers in 2-oxo acid dehydrogenase multienzyme complexes: a paradigm in the design of a multifunctional protein. Biochemistry, 1991, 30(35): 8501-8512. DOI:10.1021/bi00099a001 |
| [13] | Cronan JE. Biotin and lipoic acid: synthesis, attachment, and regulation. EcoSal Plus, 2008, 3(1): 10.1128. DOI:10.1128/ecosalplus.3.6.3.5 |
| [14] | Tezuka T, Ohnishi Y. Two Glycine riboswitches activate the Glycine cleavage system essential for Glycine detoxification in Streptomyces griseus. Journal of Bacteriology, 2014, 196(7): 1369-1376. DOI:10.1128/JB.01480-13 |
| [15] | Reed LJ, Hackert ML. Structure-function relationships in dihydrolipoamide acyltransferases. The Journal of Biological Chemistry, 1990, 265(16): 8971-8974. DOI:10.1016/S0021-9258(19)38795-2 |
| [16] | Ricaud PM, Howard MJ, Roberts EL, Broadhurst RW, Perham RN. Three-dimensional structure of the lipoyl domain from the dihydrolipoyl succinyltransferase component of the 2-oxoglutarate dehydrogenase multienzyme complex of Escherichia coli. Journal of Molecular Biology, 1996, 264(1): 179-190. DOI:10.1006/jmbi.1996.0632 |
| [17] | Zhao X, Miller JR, Cronan JE. The reaction of LipB, the octanoyl-[acyl carrier protein]: protein N-octanoyltransferase of lipoic acid synthesis, proceeds through an acyl-enzyme intermediate. Biochemistry, 2005, 44(50): 16737-16746. DOI:10.1021/bi051865y |
| [18] | Ali ST, Moir AJG, Ashton PR, Engel PC, Guest JR. Octanoylation of the lipoyl domains of the pyruvate dehydrogenase complex in a lipoyl-deficient strain of Escherichia coli. Molecular Microbiology, 1990, 4(6): 943-950. DOI:10.1111/j.1365-2958.1990.tb00667.x |
| [19] | Cao XY, Zhu L, Song XJ, Hu Z, Cronan JE. Protein moonlighting elucidates the essential human pathway catalyzing lipoic acid assembly on its cognate enzymes. Proceedings of the National Academy of Sciences USA, 2018, 115(30): E7063-E7072. DOI:10.1073/pnas.1805862115 |
| [20] | Martin N, Christensen QH, Mansilla MC, Cronan JE, de Mendoza D. A novel two-gene requirement for the octanoyltransfer reaction of Bacillus subtilis lipoic acid biosynthesis. Molecular Microbiology, 2011, 80(2): 335-349. DOI:10.1111/j.1365-2958.2011.07597.x |
| [21] | Cao XY, Hong YQ, Zhu L, Hu YY, Cronan JE. Development and retention of a primordial moonlighting pathway of protein modification in the absence of selection presents a puzzle. Proceedings of the National Academy of Sciences USA, 2018, 115(4): 647-655. DOI:10.1073/pnas.1718653115 |
| [22] | Jordan SW, Cronan JE Jr. The Escherichia coli lipB gene encodes lipoyl (octanoyl)-acyl carrier protein: protein transferase. Journal of Bacteriology, 2003, 185(5): 1582-1589. DOI:10.1128/JB.185.5.1582-1589.2003 |
| [23] | Cronan JE. The structure of lipoyl synthase, a remarkable enzyme that performs the last step of an extraordinary biosynthetic pathway. Biochemical Journal, 2014, 464(1): e1-e3. DOI:10.1042/bj20141061 |
| [24] | Morris TW, Reed KE, Cronan JE. Lipoic acid metabolism in Escherichia coli: the lplA and lipB genes define redundant pathways for ligation of lipoyl groups to apoprotein. Journal of Bacteriology, 1995, 177(1): 1-10. DOI:10.1128/jb.177.1.1-10.1995 |
| [25] | Kim DJ, Lee SJ, Kim HS, Kim KH, Lee HH, Yoon HJ, Suh SW. Structural basis of octanoic acid recognition by lipoate-protein ligase B. Proteins: Structure, Function, and Bioinformatics, 2008, 70(4): 1620-1625. |
| [26] | Harmer JE, Hiscox MJ, Dinis PC, Fox SJ, Iliopoulos A, Hussey JE, Sandy J, van Beek FT, Essex JW, Roach PL. Structures of lipoyl synthase reveal a compact active site for controlling sequential sulfur insertion reactions. The Biochemical Journal, 2014, 464(1): 123-133. DOI:10.1042/BJ20140895 |
| [27] | McLaughlin MI, Lanz ND, Goldman PJ, Lee KH, Booker SJ, Drennan CL. Crystallographic snapshots of sulfur insertion by lipoyl synthase. Proceedings of the National Academy of Sciences USA, 2016, 113(34): 9446-9450. DOI:10.1073/pnas.1602486113 |
| [28] | Lanz ND, Rectenwald JM, Wang B, Kakar ES, Laremore TN, Booker SJ, Silakov A. Characterization of a radical intermediate in lipoyl cofactor biosynthesis. Journal of the American Chemical Society, 2015, 137(41): 13216-13219. DOI:10.1021/jacs.5b04387 |
| [29] | McCarthy EL, Booker SJ. Destruction and reformation of an iron-sulfur cluster during catalysis by lipoyl synthase. Science, 2017, 358(6361): 373-377. DOI:10.1126/science.aan4574 |
| [30] | Dong G, Cao LL, Ryde U. Insight into the reaction mechanism of lipoyl synthase: a QM/MM study. JBIC Journal of Biological Inorganic Chemistry, 2018, 23(2): 221-229. DOI:10.1007/s00775-017-1522-8 |
| [31] | McCarthy EL, Booker SJ. Biochemical approaches for understanding iron-sulfur cluster regeneration in Escherichia coli lipoyl synthase during catalysis. Methods in Enzymology, 2018, 606: 217-239. |
| [32] | McCarthy EL, Rankin AN, Dill ZR, Booker SJ. The A-type domain in Escherichia coli NfuA is required for regenerating the auxiliary[4Fe-4S] cluster in Escherichia coli lipoyl synthase. Journal of Biological Chemistry, 2019, 294(5): 1609-1617. DOI:10.1074/jbc.RA118.006171 |
| [33] | Kim DJ, Kim KH, Lee HH, Lee SJ, Ha JY, Yoon HJ, Suh SW. Crystal structure of lipoate-protein ligase A bound with the activated intermediate: insights into interaction with lipoyl domains. The Journal of Biological Chemistry, 2005, 280(45): 38081-38089. DOI:10.1074/jbc.M507284200 |
| [34] | Fujiwara K, Toma S, Okamura-Ikeda K, Motokawa Y, Nakagawa A, Taniguchi H. Crystal structure of lipoate-protein ligase A from Escherichia coli: Determination of the lipoic acid-binding site. Journal of Biological Chemistry, 2005, 280(39): 33645-33651. DOI:10.1074/jbc.M505010200 |
| [35] | Cronan JE, Zhao X, Jiang YF. Function, attachment and synthesis of lipoic acid in Escherichia coli. Advances in Microbial Physiology, 2005, 50: 103-146. |
| [36] | McManus E, Luisi BF, Perham RN. Structure of a putative lipoate protein ligase from Thermoplasma acidophilum and the mechanism of target selection for post-translational modification. Journal of Molecular Biology, 2006, 356(3): 625-637. DOI:10.1016/j.jmb.2005.11.057 |
| [37] | Martin N, Lombardía E, Altabe SG, de Mendoza D, Mansilla MC. A lipA (yutB) mutant, encoding lipoic acid synthase, provides insight into the interplay between branched-chain and unsaturated fatty acid biosynthesis in Bacillus subtilis. Journal of Bacteriology, 2009, 191(24): 7447-7455. DOI:10.1128/JB.01160-09 |
| [38] | Christensen QH, Cronan JE. Lipoic acid synthesis: a new family of octanoyltransferases generally annotated as lipoate protein ligases. Biochemistry, 2010, 49(46): 10024-10036. DOI:10.1021/bi101215f |
| [39] | Christensen QH, Martin N, Mansilla MC, de Mendoza D, Cronan JE. A novel amidotransferase required for lipoic acid cofactor assembly in Bacillus subtilis. Molecular Microbiology, 2011, 80(2): 350-363. DOI:10.1111/j.1365-2958.2011.07598.x |
| [40] | Zorzoli A, Grayczyk JP, Alonzo F. Staphylococcus aureus tissue infection during Sepsis is supported by differential use of bacterial or host-derived lipoic acid. PLoS Pathogens, 2016, 12(10): e1005933. DOI:10.1371/journal.ppat.1005933 |
| [41] | Laczkovich I, Teoh WP, Flury S, Grayczyk JP, Zorzoli A, Alonzo III F. Increased flexibility in the use of exogenous lipoic acid by Staphylococcus aureus. Molecular Microbiology, 2018, 109(2): 150-168. DOI:10.1111/mmi.13970 |
| [42] | Grayczyk JP, Harvey CJ, Laczkovich I, Alonzo F III. A lipoylated metabolic protein released by Staphylococcus aureus suppresses macrophage activation. Cell Host & Microbe, 2017, 22(5): 678-687. e9. |
| [43] | Jin JQ, Hachisuka SI, Sato T, Fujiwara T, Atomi H. A structurally novel lipoyl synthase in the hyperthermophilic archaeon Thermococcus kodakarensis. Applied and Environmental Microbiology, 2020, 86(23): e01359-01320. |
| [44] | Christensen QH, Hagar JA, O'Riordan MXD, Cronan JE. A complex lipoate utilization pathway in Listeria monocytogenes. Journal of Biological Chemistry, 2011, 286(36): 31447-31456. DOI:10.1074/jbc.M111.273607 |
| [45] | Jiang YF, Cronan JE. Expression cloning and demonstration of Enterococcus faecalis lipoamidase (pyruvate dehydrogenase inactivase) as a Ser-Ser-Lys triad amidohydrolase. Journal of Biological Chemistry, 2005, 280(3): 2244-2256. DOI:10.1074/jbc.M408612200 |
| [46] | Rasetto NB, Lavatelli A, Martin N, Mansilla MC. Unravelling the lipoyl-relay of exogenous lipoate utilization in Bacillus subtilis. Molecular Microbiology, 2019, 112(1): 302-316. DOI:10.1111/mmi.14271 |
| [47] | Christensen QH, Cronan JE. The Thermoplasma acidophilum LplA-LplB complex defines a new class of bipartite lipoate-protein ligases. The Journal of Biological Chemistry, 2009, 284(32): 21317-21326. DOI:10.1074/jbc.M109.015016 |
| [48] | Posner MG, Upadhyay A, Bagby S, Hough DW, Danson MJ. A unique lipoylation system in the Archaea. FEBS Journal, 2009, 276(15): 4012-4022. DOI:10.1111/j.1742-4658.2009.07110.x |
| [49] | Cao XY, Cronan JE. The Streptomyces coelicolor lipoate-protein ligase is a circularly permuted version of the Escherichia coli enzyme composed of discrete interacting domains. The Journal of Biological Chemistry, 2015, 290(11): 7280-7290. DOI:10.1074/jbc.M114.626879 |
| [50] | Keeney KM, Stuckey JA, O'Riordan MXD. LplA1-dependent utilization of host lipoyl peptides enables Listeria cytosolic growth and virulence. Molecular Microbiology, 2007, 66(3): 758-770. DOI:10.1111/j.1365-2958.2007.05956.x |
| [51] | Grayczyk JP, Alonzo F III. Staphylococcus aureus Lipoic acid synthesis limits macrophage reactive oxygen and nitrogen species production to promote survival during infection. Infection and Immunity, 2019, 87(10): e00344-19. |
| [52] | Teoh WP, Resko ZJ, Flury S, Alonzo F III. Dynamic relay of protein-bound lipoic acid in Staphylococcus aureus. Journal of Bacteriology, 2019, 201(22): e00446-19. |
| [53] | Zhu KM, Chen H, Jin J, Wang N, Ma GX, Huang JD, Feng YJ, Xin JQ, Zhang HM, Liu HG. Functional identification and structural analysis of a new lipoate protein ligase in Mycoplasma hyopneumoniae. Frontiers in Cellular and Infection Microbiology, 2020, 10: 156. |
| [54] | Ma Q, Zhao X, Eddine AN, Geerlof A, Li X, Cronan JE, Kaufmann SHE, Wilmanns M. The Mycobacterium tuberculosis LipB enzyme functions as a cysteine/lysine dyad acyltransferase. Proceedings of the National Academy of Sciences USA, 2006, 103(23): 8662-8667. |
| [55] |
Ma JR, Liu AN, Zhang WB, Mao YH, Yu YH. LipB-LipA is the only lipoyl acylation pathway in Xanthomonas campestris. Acta Microbiologica Sinica, 2020, 60(8): 1729-1740.
(in Chinese) 马建荣, 刘安娜, 张文彬, 毛雅慧, 余永红. 野油菜黄单胞菌中LipB-LipA是唯一的硫辛酰化途径. 微生物学报, 2020, 60(8): 1729-1740. |
| [56] | 陶乃敏. 硫辛酸合成的工艺研究. 天津大学硕士学位论文, 2014. |
| [57] | Sun YR, Zhang WB, Ma JC, Pang HS, Wang HH. Overproduction of α-lipoic acid by gene manipulated Escherichia coli. PLoS ONE, 2017, 12(1): e0169369. |
 2021, Vol. 61
2021, Vol. 61