
中国科学院微生物研究所,中国微生物学会
文章信息
- 牛晓倩, 陶楠, 舒泉先, 杨小雁, 从瑞涛, 周胜虎, 邓禹. 2022
- NIU Xiaoqian, TAO Nan, SHU Quanxian, YANG Xiaoyan, CONG Ruitao, ZHOU Shenghu, DENG Yu.
- 代谢工程强化脱氮副球菌DYTN-1去除氮素污染物
- Metabolic engineering enhances the nitrogen removal by Paracoccus denitrificans DYTN-1
- 微生物学报, 62(10): 3932-3946
- Acta Microbiologica Sinica, 62(10): 3932-3946
-
文章历史
- 收稿日期:2022-02-22
- 修回日期:2022-04-18
- 网络出版日期:2022-06-09
2. 江南大学江苏省生物活性制品加工工程技术研究中心, 江苏 无锡 214122;
3. 山东渤海油脂工业有限公司, 山东 滨州 256599;
4. 帝斯曼江山制药(江苏)有限公司, 江苏 靖江 214500
2. Jiangsu Provincial Research Center for Bioactive Product Processing Technology, Jiangnan University, Wuxi 214122, Jiangsu, China;
3. Shandong Bohi Oils & Fats Industry Co., Ltd., Binzhou 256599, Shandong, China;
4. DSM Jiangshan Pharmaceutical (Jiangsu) Company, Jingjiang 214500, Jiangsu, China
随着人类活动的日益频繁,高氮素含量的污水不断排入河流、湖泊和其他水体中,导致水质富营养化,不仅使环境生态失衡,而且产生饮用水安全问题[1‒2]。因此,为了应对这些挑战,氮素必须在污水系统中被快速脱除[3]。目前,生物脱氮是最常用和最有效的处理方法[4],而选择具有不同脱氮功能的菌株对复杂的污水治理很重要。然而,自然界存在的菌株在污水环境中通常生长速度较慢[5],不能满足工业脱氮的一般要求。以环境微生物为底盘细胞,利用合成生物技术和代谢工程强化脱氮功能,是突破上述挑战的重要方法。
目前,从活性污泥中分离出的几种环境友好型细菌,如假单胞菌和芽孢杆菌[6],已被用作底盘来降解石油[7]、农药污染物[8]和富营养化污染物[9‒10]。在工程化改造这些微生物的过程中,标准的遗传操作方法和合成生物学工具,例如启动子、终止子、质粒和选择标记已得到广泛开发[11‒13]。使用基于sacB的基因组修饰系统(包括pK18mobsacB和pBBR1MCS3质粒),Chen等成功整合并敲除了从活性污泥中分离出的Comamonas testosterone CNB-1菌株中的基因,并鉴定了受MarR型调节因子(GenR)调控的苯甲酰辅酶A和3-羟基苯甲酸诱导型启动子PgenA[14]。基于这些基因表达系统,上述环境友好型细菌可以作为脱氮途径基因表达的理想底盘系统。
与不具有脱氮功能的菌株相比,使用具有脱氮功能的菌株具有更多的优势。主要是因为脱氮微生物自身具有丰富的脱氮酶系,具有更好的脱氮效果。然而,以脱氮微生物为底盘细胞,通过代谢工程强化脱氮功能的研究却鲜有报道。从活性污泥中分离的脱氮副球菌(Paracoccus denitrificans)是一种环境友好的α-变形菌纲菌株,属于典型的异养硝化好氧反硝化菌株(heterotrophic nitrifying and aerobic denitrifying bacteria,HN-AD),其打破了传统反硝化必须在厌氧条件下进行的界限。在有氧条件下,好氧反硝化菌可同时利用环境中的O2和NO3−或者在进行反硝化过程中产生的中间代谢物(如NO和N2O)作为反应所需的电子受体,进行反硝化反应,最终生成气态氮化合物并同时去除污水中的COD,具有较好的脱氮能力,是代谢工程改造的理想底盘[15‒16]。代谢工程改造首先需要遗传操作工具,如表达质粒和启动子等。目前关于P. denitrificans的遗传操作工具报道较少。2009年,Ind等发现质粒pIND4可在P. denitrificans中表达[17],其主要包含pMG170骨架[18]、lacIq[19]和来自pJBA24的IPTG诱导型启动子[20],经异丙基-β-d-硫代半乳糖苷(isopropyl-β-d-thiogalactoside,IPTG)诱导后,在pIND4上过表达的CheY6蛋白表达水平比未诱导菌株高约3倍[17]。然而,这种较低的倍数并不能满足代谢工程的各种要求。Kaczmarczyk等利用组成型合成启动子cumate响应性的CymR阻遏蛋白和CymR操纵位点,开发了一种cumate诱导型基因表达系统,从而实现了4 700倍的诱导变化[21]。此外,脱氮副球菌属于α-变形菌纲,Tett等在α-变形菌纲中建立了具有不同拷贝数的质粒pLMB51 (低拷贝)和pLMB509 (高拷贝)及其牛磺酸诱导表达启动子PtauA,为脱氮副球菌中的基因表达提供了工具[22]。尽管在以前的研究中取得了重大突破,但目前仍然缺乏可在P. denitrificans中应用的氮素诱导型启动子或组成型启动子,这限制了代谢工程改造的实施。
本研究室从活性污泥中分离出1株脱氮副球菌P. denitrificans DYTN-1,具有良好的脱氮效果[23]。在本研究中,测定了P. denitrificans DYTN-1的全基因组序列,并挖掘出了6个内源性启动子,可被NO2‒、NO3‒和NH4+诱导。随后,以P. denitrificans DYTN-1为底盘细胞,使用代谢工程方法强化了氮素降解功能。
1 材料与方法 1.1 菌株和菌株的培养大肠杆菌(E. coli) JM109、P. denitrificans DYTN-1和E. coli S17-1λpir分别作为质粒克隆宿主、基因表达宿主和接合转移的辅助菌。LB培养基用于培养大肠杆菌和P. denitrificans DYTN-1,模拟污水培养基[24] [10 g/L甘油、0.5 g/L NaCl、6.78 g/L Na2HPO4、3 g/L KH2PO4、1 g/L NH4Cl (或1 g/L NaNO2或2 g/L KNO3)、0.24 g/L MgSO4 7H2O,11.1 mg/L CaCl2]用于P. denitrificans DYTN-1的氮素降解测定。根据需要向培养基中加入氨苄青霉素(100 μg/mL)、壮观霉素(50 μg/mL)、四环素(50 μg/mL)、氯霉素(50 μg/mL)和卡那霉素(50 μg/mL)。氮素测定时,将8 mL P. denitrificans DYTN-1培养过夜的种子液加入含有200 mL模拟污水培养基的锥形瓶(500 mL)中,并在30 ℃和150 r/min条件下培养,每隔12 h进行取样,检测菌体生长情况以及氮源消耗情况。其中,以野生菌株作为对照,携带重组质粒pKCE-P1851-amoA、pKCE-P1851-amoB和pKCE-P1851-hao的P. denitrificans DYTN-1在以NH4Cl (1 g/L)为氮源的模拟污水培养基中培养,携带重组质粒pKCE-P1746-nirS的P. denitrificans DYTN-1在以KNO3 (2 g/L)为氮源的培养基中生长。携带重组质粒pKCE-P1851-hao-P1746-nirS的P. denitrificans DYTN-1在以NH4Cl (1 g/L)和KNO3 (2 g/L)为氮源的模拟污水培养基中培养。
1.2 接合试验表达重组质粒的E. coli S17-1λpir作为供体菌株,P. denitrificans DYTN-1作为受体菌株。将E. coli S17-1λpir和P. denitrificans DYTN-1分别接种到含有5 mL LB培养基的试管中,并在37 ℃和30 ℃下培养过夜。第2天,分别5 000 r/min离心2 min收集108 CFU/mL细胞,并用LB培养清洗2次,再使用3:10的比例(E. coli S17-1λpir: P. denitrificans DYTN-1)将细胞转移到同一管中混合,37 ℃下培养30 min。随后,剧烈摇晃试管以破坏供体菌和受体菌之间的性菌毛连接,从而停止基因转移。最后,将0.1 mL的接合菌液涂布在壮观霉素和卡那霉素的双抗琼脂平板上筛选阳性克隆。
1.3 基因组测序、组装与注释P. denitrificans DYTN-1基因组测序由苏州金唯智生物科技有限公司完成。采用全基因组扩增策略构建不同插入片段的文库,使用Qubit 3.0和Agilent 2100对文库质量进行检测。检测合格后基于PacBio测序平台对这些文库进行测序,采用HGAP软件和Falcon软件对纯三代数据进行组装。采用Prodigal软件对细菌基因组进行基因预测。编码蛋白质的基因通过与NR (Non-redundant)数据库、KEGG (Kyoto Encyclopedia of Genes and Genomes)、COG (Cluster of Orthologous Groups of Proteins)数据库和GO (Gene Ontology)数据库进行BLAST比对,筛选条件为e-value≤1e‒5,获取比对得分最高的结果,完成蛋白序列功能注释。
1.4 质粒构建DNA聚合酶和限制酶购自TaKaRa。使用质粒pJKR-H-cdaR作为模板[25],使用引物对sfgfp-F/sfgfp-R对绿色荧光蛋白(sfGFP)基因碱基序列进行PCR扩增,并克隆到pIND4的Nco Ⅰ/Hind Ⅲ位点,生成pIND4-sfgfp质粒。然后,使用P. denitrificans DYTN-1基因组DNA作为模板,内源启动子PrpsU、P0893、P1746、P1845、P1851和P4130通过引物对rpsu-F/rpsu-R、0893-F/0893-R、1746-F/1746-R、1845-F/1845-R、1851-F/1851-R和4130-F/4130-R扩增获得,这些诱导型启动子被克隆到pIND4-sfgfp中sfgfp的上游以替换PA1/04/03,产生以下质粒:pKCE-PrpsU-sfgfp、pKCE-P0893-sfgfp、pKCE-P1746-sfgfp、pKCE-P1845-sfgfp、pKCE-pKCE-P1851-sfgfp和pKCE-P4130-sfgfp。
使用引物对nirS-F/nirS-R从P. denitrificans DYTN-1基因组DNA中扩增nirS基因。分别合成来自Nitrosomonas europaea、Nitrosomonas europaea和Nitrosomonas sp. ENI-11的amoA、amoB和hao基因。将amoA、amoB、hao和nirS的CDS序列插入pKCE-P1851-sfgfp、pKCE-P1851-sfgfp、pKCE-P1851-sfgfp和pKCE-P1746-sfgfp的Nco Ⅰ/Hind Ⅲ位点,分别生成pKCE-P1851-amoA、pKCE-P1851-amoB、pKCE-P1851-hao和pKCE-P1746-nirS质粒。使用引物对1746nirS-F/ 1746nirS-R从pKCE-P1746-nirS扩增包含nirS基因和调控其表达的启动子P1746,再以pKCE-P1851-hao为质粒载体通过同源重组方式得到质粒pKCE-P1851-hao-P1746-nirS。所有菌株、质粒和引物序列见表 1和表 2。
| Name | Properties | Sources |
| Strains | ||
| P. denitrificans DYTN-1 | Wild type, for gene expression | Lab preserved |
| E. coli JM109 | Wild type, for plasmid construction | Lab preserved |
| E. coli S17-1λpir | Wild type, plasmid transfer helper strain | Lab preserved |
| Plasmids | ||
| pIND4 | ColE1 ori, oriT, repA, KanR | Lab preserved |
| pIND4-sfgfp | pIND4 carrying the gene sfgfp, KanR | This study |
| pKCE-PrpsU-sfgfp | pIND4 backbone, PrpsU controlling the expression of sfgfp, KanR | This study |
| pKCE-P0893-sfgfp | pIND4 backbone, P0893 controlling the expression of sfgfp, KanR | This study |
| pKCE-P1746-sfgfp | pIND4 backbone, P1746 controlling the expression of sfgfp, KanR | This study |
| pKCE-P1845-sfgfp | pIND4 backbone, P1845 controlling the expression of sfgfp, KanR | This study |
| pKCE-P1851-sfgfp | pIND4 backbone, P1851 controlling the expression of sfgfp, KanR | This study |
| pKCE-P4130-sfgfp | pIND4 backbone, P4130 controlling the expression of sfgfp, KanR | This study |
| pKCE-P1851-amoA | pIND4 backbone, P1851 controlling the expression of amoA, KanR | This study |
| pKCE-P1851-amoB | pIND4 backbone, P1851 controlling the expression of amoB, KanR | This study |
| pKCE-P1851-hao | pIND4 backbone, P1851 controlling the expression of hao, KanR | This study |
| pKCE-P1746-nirS | pIND4 backbone, P1746 controlling the expression of nirS, KanR | This study |
| pKCE-P1851-hao-P1746-nirS | pIND4 backbone, P1851 controlling the expression of hao, P1746 controlling the expression of nirS, KanR | This study |
| Primers | Sequences (5′→3′) | Restriction sites |
| sfgfp-F | CCATGGTGCGTATAGGTGAAGAACTG | Nco I |
| sfgfp-R | AAGCTTAGAACTGGCATGCATC | Hind Ⅲ |
| rpsu-F | GAGCTTCATTTCACGGAACACCCC | |
| rpsu-R | GGCACTCTCCTCATTCTTCATGCC | |
| 0893-F | GATTTCCTCGGCCGCAGGC | |
| 0893-R | GACCGGCAGAGCGGACAG | |
| 1746-F | GTCCAGCGACAGCTCGCC | |
| 1746-R | GTCGGTCGTCCTTTTCCTTGTTCC | |
| 1851-F | CATCCTCGAGCTGCTGGG | |
| 1851-R | ACTGCTATCCTGCCGCGG | |
| 4130-F | TGGCGCAGATCGCGCATG | |
| 4130-R | GAAGGCACCTCGGTCGCC | |
| nirS-F | ATGAGACAAAGGACCCCATTCG | |
| nirS-R | TCAATAGGTGTCGGTCATAGTGTTG | |
| 1746nirS-F | GTCCAGCGACAGCTCGCC | |
| 1746nirS-R | TCAATAGGTGTCGGTCATAGTGTTG |
1.5 氮素的定量检测
硝酸盐采用百里酚分光光度法(GB/T5750.5—2006),氨氮采用纳氏试剂分光光度法(HJ 535—2009),总氮采用碱性过硫酸钾消解紫外分光光度法(HJ 636—2012)。其中,为保证数据的准确性,菌液需经5 000 r/min离心6 min后取上清液进行硝酸盐和氨氮的测定。总氮测定时则不需离心处理,直接对含有菌体和培养基的样品进行测定,以同时测定被菌体同化吸收的氮素和培养基中残余氮素。同时,氮素的测定均采用标准品进行标准曲线的绘制,再根据标准曲线计算样品氮素的含量。测量时使用试剂盒进行硝酸盐(连华LH-NO3)、氨氮(连华LH-NH4)和总氮(连华LH-NT)检测。所有测试均严格按照制造商的说明进行。
1.6 诱导型启动子的筛选和表征将含有需要表征启动子的菌株P. denitrificans DYTN-1过夜活化后,以2%的接种量接种至含2 mL模拟污水培养基(4 g/L的甘油作为碳源)的48孔板中培养。在接种的同时分别添加不同浓度的氮源(蛋白胨、铵盐、亚硝酸盐和硝酸盐)进行菌株培养并诱导启动子表达,诱导培养48 h后取出菌液并用10 mmol/L磷酸盐缓冲液(pH 7.0)稀释,以确保OD600在0.2‒0.8范围内。在室温下使用BioTek HT酶标仪(Winooski)在(485±20) nm激发波长和(528±20) nm发射波长下测定荧光强度。以荧光强度和OD600的比值表征启动子强度。
2 结果与分析 2.1 P. denitrificans DYTN-1来源的氮素诱导型内源启动子筛选作为代谢工程的重要元件,启动子具有调节基因表达的关键作用[26]。通常,化合物分解代谢途径的基因表达受到分解代谢底物或中间产物诱导[27‒28]。为了了解氮素降解相关代谢途径和功能,挖掘受氮素诱导的启动子用于后续代谢工程调控,我们对P. denitrificans DYTN-1进行了全基因组测序。分析发现,共有3 765个基因被分配到6个主要的KEGG途径和40个子途径中(图 1)。代谢是6种主要KEGG途径中最大的一类。在所有子途径中,碳水化合物代谢(489条基因,占比12.99%)是最大的类别,其次是氨基酸代谢(471条基因,占比12.51%)、全局和概览图(407条基因,占比10.81%)、膜转运(349条基因,占比9.27%)、能量代谢(270条基因,占比8.50%)和细胞群落—原核生物(189条基因,占比5.02%)。P. denitrificans DYTN-1的基因功能注释为探索基因组研究中基因产物的特定生物过程、功能、亚细胞定位和途径提供了基础。
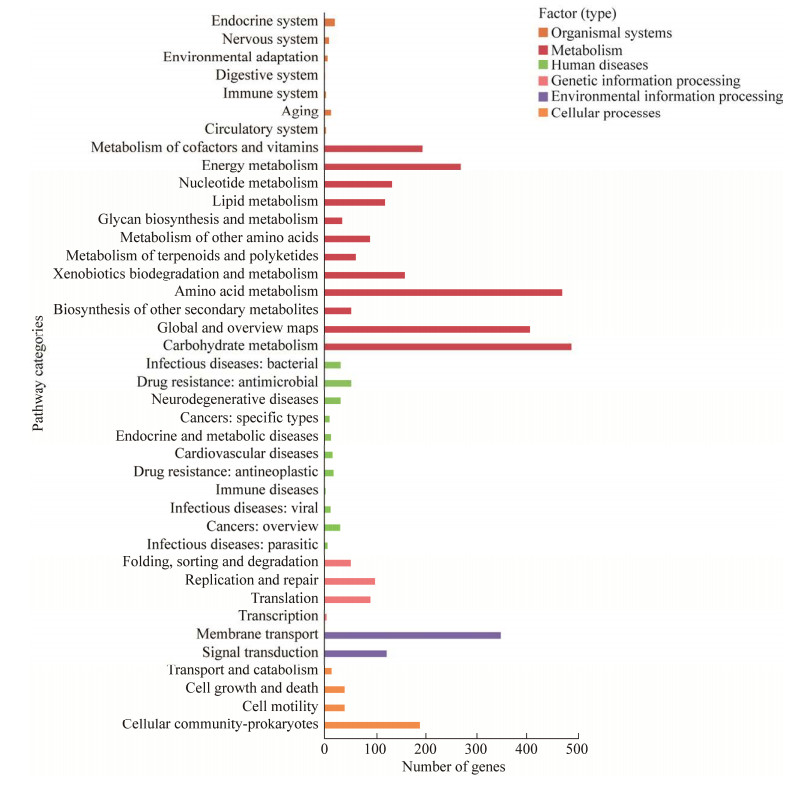
|
| 图 1 脱氮副球菌DYTN-1基因的KEGG通路富集分析 Figure 1 KEGG pathway enrichment analysis for the Paracoccus denitrificans DYTN-1 genes. A total of 3 765 genes were assigned to 6 major KEGG pathways and 40 sub-pathways. |
根据Giannopoulos等[29]的报道,泛醌生物合成蛋白COQ9、粪卟啉原Ⅲ氧化酶、细胞色素c过氧化物酶、核苷二磷酸激酶、细胞色素c氧化酶cbb3型亚基Ⅲ和信号转导组氨酸激酶与氮素降解代谢密切相关。通过氨基酸比对发现,P. denitrificans DYTN-1基因组中基因rpsU、1851、0893、1746、1845和4130与泛醌生物合成蛋白COQ9、粪卟啉原Ⅲ氧化酶、细胞色素c过氧化物酶、核苷二磷酸激酶、细胞色素c氧化酶cbb3型亚基Ⅲ和信号转导组氨酸激酶同源性高达100%。因此,这6个基因上游启动子可能是受氮素诱导的启动子。选取基因CDS上游400 bp序列作为候选启动子[29],选定的启动子序列包括核心启动子区域(‒10区和‒35区)、5′-非翻译区(5ʹ untranslated region,5ʹ-UTRs)和核糖体结合位点(ribosome bind site,RBS)区域,可直接克隆到靶基因CDS区域进行表达。
在P. denitrificans DYTN-1中,利用NH4+、NO2‒和NO3‒作为诱导剂诱导6种启动子启动sfGFP蛋白的表达,蛋白胨作为阴性对照,研究所选启动子对氮素的诱导性能。发现蛋白胨诱导后启动子启动的sfGFP蛋白表达水平非常低,而在无机氮存在下sfGFP表达水平较高。具体而言,P0893对NO2‒、NO3‒和NH4+的有效响应范围分别为250‒2 000 mg/L、750‒2 000 mg/L和500‒750 mg/L (图 2A)。NO2‒诱导产生15倍的表达差异,并且在NO2‒浓度和诱导强度之间表现出线性关系,然而对NO3‒和NH4+的反应相对较低。P1851对NO3‒和NH4+的有效响应范围为250‒2 000 mg/L和500‒2 000 mg/L,分别产生9倍和13倍的诱导变化(图 2B)。P1746对NO3‒和NH4+的有效响应范围分别为250‒2 000 mg/L和100‒2 000 mg/L,NO2‒只产生很低水平的表达差异。NO3‒和NH4+的诱导分别产生了19倍和16倍的表达差异,NO3‒浓度和诱导强度之间存在线性关系(图 2C)。P4130对NO2‒、NO3‒和NH4+的有效响应范围分别为50‒2 000 mg/L、50‒2 000 mg/L和75‒2 000 mg/L。其中,NH4+浓度在750‒2 000 mg/L之间的表达差异明显提高,最终达到最高表达差异11倍(图 2D)。P1845对NO2‒、NO3‒和NH4+的有效响应范围分别为50‒2 000 mg/L、50‒1 000 mg/L和75‒1 000 mg/L。NO2‒和NO3‒的诱导分别产生11和15倍的诱导变化(图 2E)。PrpsU对NO2‒、NO3‒和NH4+的有效响应范围分别为50‒2 000 mg/L、50‒1 000 mg/L和50‒2 000 mg/L。其中,低浓度的NH4+ (50‒2 000 mg/L)明显提高了诱导强度,表现出NH4+浓度与表达差异的线性关系,最终在2 000 mg/L时达到最高的表达差异,为26倍(图 2F)。PrpsU的有效响应下限为50 mg/L,远低于P1851、P0893、P1746、P1845和P4130。因此,PrpsU启动子具有通过监测氨氮浓度变化来动态调节脱氮基因表达的潜力。
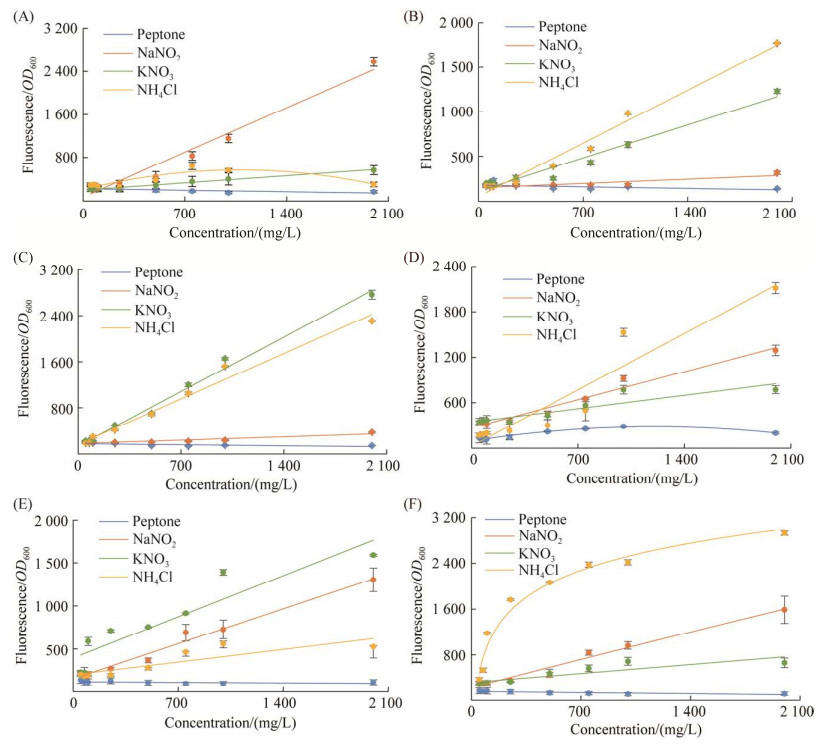
|
| 图 2 不同启动子受NO2‒、NO3‒和NH4+诱导的剂量曲线 Figure 2 Induction dose curves of different promoters induced by NO2‒, NO3‒ and NH4+. A–F: induction dose curves for P0893 (A), P1851 (B), P1746 (C), P4130 (D), P1845 (E), and PrpsU (F) using different nitrogen induction sources. Peptone did not induce promoter expression and was used as a control. The inducer concentration range that could generate 1.5-fold to highest of induction fold change was defined as the effective response range. Data represents the mean and standard deviation for three replicates. |
2.2 以P. denitrificans DYTN-1为底盘的代谢工程改造可行性研究
P. denitrificans DYTN-1是从活性污泥中分离出来的天然菌株[23],其基因操作特性依然未知。为探究P. denitrificans DYTN-1作为底盘细胞的可行性,首先研究了它的抗生素耐受性,以确认接合过程的阳性克隆筛选标记。结果发现,P. denitrificans DYTN-1对壮观霉素耐受,但对氨苄青霉素、卡那霉素、氯霉素和四环素不耐受。相比之下,E. coli S17-1λpir对所有测试的抗生素都敏感(图 3A)。同时,P. denitrificans DYTN-1的生长几乎不受壮观霉素浓度的影响,而E. coli S17-1λpir的生长受壮观霉素影响较大,在壮观霉素为30 µg/mL时可完全抑制E. coli S17-1λpir的生长(图 3B‒C)。因此,使用壮观霉素搭配氯霉素、卡那霉素、四环素或氨苄霉素,制备双抗生素琼脂平板,用于在接合后筛选阳性P. denitrificans DYTN-1克隆。接合研究表明,pIND4[17]成功转移到P. denitrificans DYTN-1中并产生了较高的接合效率(图 3D)和70%的阳性率(图 3E)。
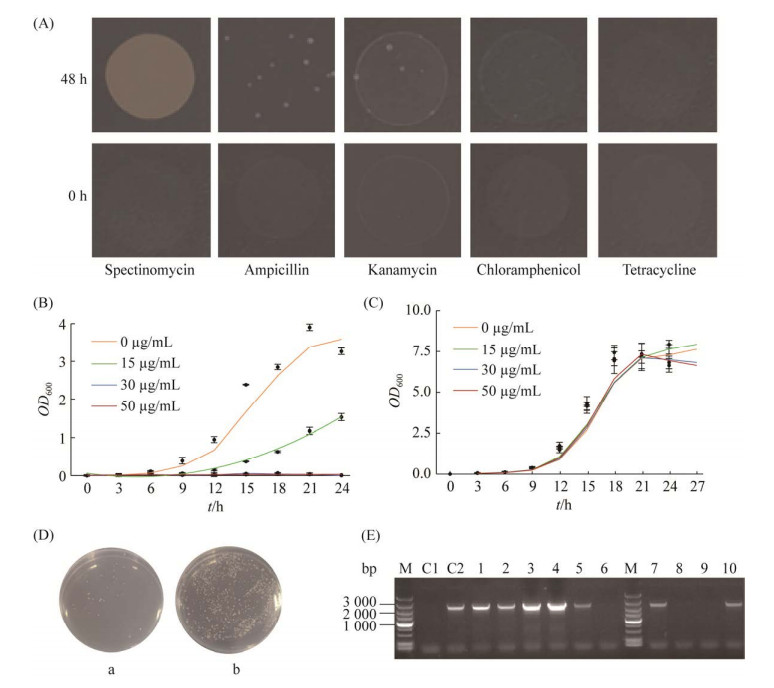
|
| 图 3 验证P. denitrificans DYTN-1接合的可行性 Figure 3 Testing P. denitrificans DYTN-1 conjugation feasibility. A: P. denitrificans DYTN-1 growth on different antibiotic plates. B: growth of E. coli S17-1λpir in media containing different concentrations of spectinomycin. Data represents the mean and standard deviation for three replicates. C: growth of P. denitrificans DYTN-1 in media containing different concentrations of spectinomycin. Data represents the mean and standard deviation for three replicates. D: the conjugative transformation efficiency of P. denitrificans DYTN-1. Empty and pIND4 containing E. coli S17-1λpir cells were used for conjugative transformation. The dense colonies on the agar plate (right) show a high conjugation efficiency. a: P. denitrificans DYTN-1 growth on double antibiotic antibacterial plates (spectinomycin 30 µg/mL and kanamycin 50 µg/mL), b: P. denitrificans-pIND4 growth on double antibiotic antibacterial plates. P. denitrificans- pIND4: P. denitrificans DYTN-1 strain carrying plasmid pIND4. E: assessment the positive rate of conjugation transformation by colony PCR. M: marker. C1: negative control. C2: positive control. Target band is 2 485 bp. After colony PCR, we found 7 positive colonies in 10 candidates, thus indicating the positive rate is 70% (7/10). |
异源质粒的遗传稳定性是另一个评估遗传可操作性的关键指标。利用P. denitrificans DYTN-1的内源启动子PrpsU控制pIND4质粒中sfGFP蛋白的表达,并构建了pKCE-PrpsU-sfgfp质粒。结果发现,pKCE-PrpsU-sfgfp质粒在10轮传代培养中保持高水平的稳定性(图 4)。根据荧光分析,pKCE-PrpsU-sfgfp在P. denitrificans DYTN-1中的荧光值离散系数稳定在0.12±0.02。脱氮副球菌DYTN-1中稳定的sfGFP表达表明该质粒系统可在菌株中稳定复制和遗传表达,从而表现出良好的遗传稳定性。总体而言,上述研究表明P. denitrificans DYTN-1具有作为代谢工程底盘细胞强化氮素降解的潜力。
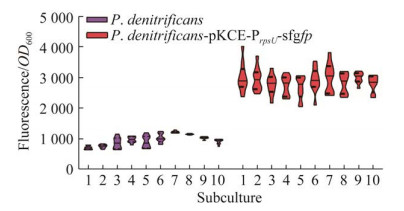
|
| 图 4 外源质粒的稳定性分析 Figure 4 Stability analysis of heterogeneous plasmids. The coefficient of dispersion (c) was the ratio of the standard deviation (σ) of a set of data to its corresponding mean (µ). This is a relative indicator for measuring the degree of data dispersion. It is mainly used to compare the degree of dispersion of different groups of data. c=σ/µ. Each violin plot contains 10 parallel data. |
2.3 代谢工程强化P. denitrificans DYTN-1脱氮
氨单加氧酶(AMO)、羟胺氧化酶酶(HAO)、硝酸盐还原酶(NAR)、亚硝酸盐还原酶(NIRS)、NO还原酶和N2O还原酶是氨氮经硝化和反硝化作用转化为氮气的催化酶(图 5A)[30]。其中,AMO和HAO分别在催化NH4+转化为NH2OH和NH2OH转化为NO2‒的硝化作用中起着不可或缺的作用。此外,NirS负责将NO2‒还原为NO。由于NAR的电子亲和力高于NirS,在反硝化过程中电子优先传递给NAR从而还原NO3‒,NO2‒还原受到NirS活性的限制。因此,NirS是反硝化过程的限速步骤[31]。为了强化P. denitrificans DYTN-1的脱氮功能,以pKCE-P1851-sfgfp为载体过表达amoA、amoB和hao,以pKCE-P1746-sfgfp为载体表达nirS。
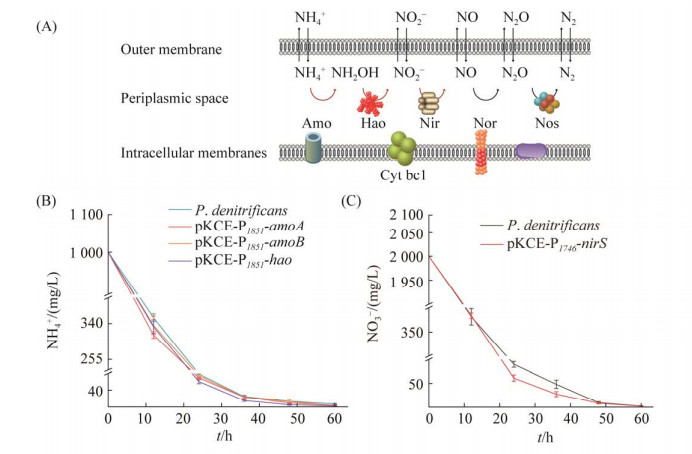
|
| 图 5 代谢工程强化P. denitrificans DYTN-1硝化和反硝化途径基因表达 Figure 5 Metabolic engineering enhances nitrification and denitrification pathway gene expression in P. denitrificans DYTN-1. A: metabolic engineering enhances the nitrogen metabolism pathway. NOR: nitric oxide reductase; NOS: nitrous oxide reductase; Cyt bc1: cytochrome bc1. Red arrows represent overexpressed genes. B: changes in NH4+ during treatment. Data represents the mean and standard deviation for three replicates. C: NO3‒ changes during treatment. Data represents the mean and standard deviation for three replicates. |
为了构建高效脱氮菌株并加速脱氮过程,分别选择受NH4+和NO3‒诱导型中等强度启动子P1851和P1746来控制amoA、amoB、hao和nirS的表达。从而当环境中NH4+浓度过高时诱导启动硝化途径的基因表达,在NO3‒积累过高时诱导启动反硝化途径基因表达,即可避免在没有NH4+或NO3‒污染时的基因过量表达给细胞造成的代谢负担,也可实现脱氮副球菌对氮素污染的实时监控和强化降解。实验数据表明,在处理的前20 h,工程菌株的氨氮去除率高于野生型(图 5B)。处理20 h后,残留氨氮水平约为250 mg/L,低于启动子对NH4+的有效响应下限,对启动子P1851诱导作用减弱,将会导致amoA、amoB和hao表达水平降低。因此,20 h后amoA、amoB和hao过表达与野生型菌株的脱氮率相似。最终,用过表达hao的工程菌处理后,培养基中NH4+残留了2.4 mg/L,是野生型菌株的35%。
我们还利用NO3‒诱导型启动子P1746过表达nirS来增强反硝化作用。处理24 h后,nirS过表达菌株中培养基中NO3‒的残余量为野生型菌株的67% (图 5C)。处理36 h后,NO3‒残留量约为25 mg/L,低于P1746启动子对NO3‒的有效响应下限,从而导致质粒上的nirS基因表达关闭,仅有基因组的原生nirS基因表达,工程菌株的反硝化速率降低到与野生菌相似。最终,在处理60 h后,nirS过表达的工程菌株培养基中的NO3‒残余量为1.9 mg/L。
在硝化反应进程中过表达hao的工程菌促进NH4+降解效果较好。因此,为了进一步加强氮素降解,构建了质粒pKCE-P1851-hao-P1746-nirS,在P. denitrificans DYTN-1中同时过表达hao和nirS基因,以达到同时促进硝化和反硝化反应的目的。处理12 h后,培养基中NH4+和NO3‒的残余率分别为41.2%和11.5%,其中NO3‒残留量为229.74 mg/L,是野生菌株的一半,且远小于仅单过表达nirS的工程菌处理12 h后NO3‒的剩余量(图 6A‒B)。最终,同时过表达hao和nirS基因的重组菌株处理60 h后总氮(包含被细胞同化的氮素)残留了113.9 mg/L,是野生菌株的51%,处理前后的总氮减少了437.9 mg/L,降解效率达79.5% (图 6C)。培养基中NH4+和NO3‒分别残余83.8 mg/L和8.4 mg/L。
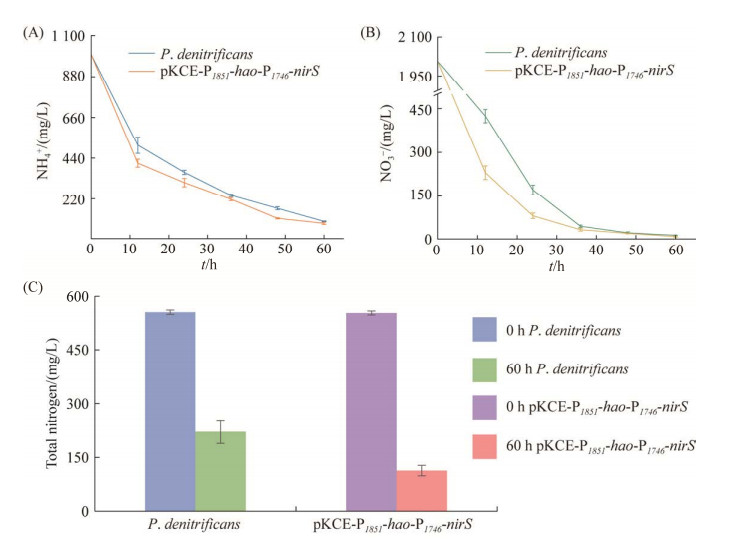
|
| 图 6 同时过表达hao和nirS强化P. denitrificansDYTN-1的脱氮能力 Figure 6 Simultaneous overexpression of hao and nirS genes enhanced the nitrogen removal ability of P. denitrificans DYTN-1. A: changes in NH4+ during treatment. B: NO3‒ changes during treatment. C: residual total nitrogen levels at 60 h of treatment in the hao and nirS overexpressed strain. Data represents the mean and standard deviation for three replicates. pKCE-P1851-hao-P1746-nirS: P. denitrificans DYTN-1 carrying plasmid pKCE-P1851-hao-P1746-nirS. |
通过上述强化硝化和反硝化途径发现NO3‒比NH4+更容易降解,这可能与amoA、amoB和hao均来自自养硝化菌有关。由于P. denitrifican是一类经典的异养硝化好氧反硝化菌,在基因组中尚未发现与自养硝化菌的amoA、amoB和hao同源的基因,而amoA需要amoB等基因的协同以及电子传递链的配合才能行使氨氧化功能。因此单独过表达amoA或amoB对NH4+的降解强化效果有限。
3 讨论与结论P. denitrificans是研究好氧反硝化的模型菌株,其优越的脱氮性能备受研究者们关注。然而,由于遗传操作困难和缺乏遗传操作工具,导致以P. denitrificans为底盘进行代谢工程改造的报道较少。本研究应用P. denitrificans DYTN-1作为底盘细胞,挖掘出6个不同氮素诱导的内源性启动子,并验证了遗传操作的可行性。最后。成功地强化了P. denitrificans DYTN-1的硝化和反硝化过程,脱氮能力显著增强。
目前,很少有研究构建脱氮副球菌的表达系统。基于pIND4和PQ5[21]质粒的IPTG和cumate诱导系统是目前2种可用的表达系统。pIND4质粒来源于Rhodobacter blasticus内源质粒pMG160,主要包括mobS、mobL和repA,mobS和mobL与接合质粒的细胞间穿梭有关,repA与质粒复制密切相关。pIND4的IPTG诱导仅产生3倍的诱导变化,不利于基因高强度表达。基于PQ5的质粒具有显著的高诱导变化,但外源添加诱导剂无疑会增加环境的负担。本研究发现,pIND4具有较高的稳定性,然而这些质粒较大(7 500‒10 900 bp),限制了多个基因在同一质粒上的表达。因此,进一步进行质粒结构简化将有助于未来的应用。
NH4+、NO2‒和NO3‒是水体中的主要氮污染物[32]。因此,筛选响应这些无机盐的启动子对于优化脱氮相关基因的表达至关重要,可以用于设计和构建复杂的动态脱氮途径[33]。具体来说,当氮污染物在环境中积累过多时,NH4+、NO2‒和NO3‒诱导型启动子开始表达,从而启动硝化或反硝化途径基因表达,强化氮素降解功能。当没有氮污染物积累时,可以降低表达水平以节省细胞的物质和能量。
目前,P. denitrificans的研究主要集中在生理、生化和功能研究上。使用以P. denitrificans为底盘的代谢工程改造研究鲜有报道。本研究利用内源性启动子在氮素诱导条件下促进了硝化和反硝化过程,为进一步挖掘基因调控工具和强化氮素降解提供了方法和思路,也为环境废水中的基因工程菌株应用提供了参考。
| [1] | Eneji AE. Review of current status and research approaches to nitrogen pollution in farmlands. Agricultural Sciences in China, 2009, 8(7): 843-849. DOI:10.1016/S1671-2927(08)60286-2 |
| [2] | Schullehner J, Hansen B, Thygesen M, Pedersen CB, Sigsgaard T. Nitrate in drinking water and colorectal cancer risk: a nationwide population-based cohort study. International Journal of Cancer, 2018, 143(1): 73-79. DOI:10.1002/ijc.31306 |
| [3] | Zuo R, Chen XJ, Li XB, Shan D, Yang J, Wang JS, Teng YG. Distribution, genesis, and pollution risk of ammonium nitrogen in groundwater in an arid loess plain, northwestern China. Environmental Earth Sciences, 2017, 76(17): 1-16. |
| [4] | Ghafari S, Hasan M, Aroua MK. Bio-electrochemical removal of nitrate from water and wastewater—a review. Bioresource Technology, 2008, 99(10): 3965-3974. DOI:10.1016/j.biortech.2007.05.026 |
| [5] | Wang SJ, Loh KC. New cell growth pattern on mixed substrates and substrate utilization in cometabolic transformation of 4-chlorophenol. Water Research, 2000, 34(15): 3786-3794. DOI:10.1016/S0043-1354(00)00144-5 |
| [6] | Sun XW, Chen L, Liu C, Xu Y, Ma W, Ni H. Biodegradation of CP/TCP by a constructed microbial consortium after comparative bacterial community analysis of long-term CP domesticated activated sludge. Journal of Environmental Science and Health, Part B, 2020, 55(10): 898-908. DOI:10.1080/03601234.2020.1794453 |
| [7] | Fulekar MH. Microbial degradation of petrochemical waste-polycyclic aromatic hydrocarbons. Bioresources and Bioprocessing, 2017, 4(1): 28. DOI:10.1186/s40643-017-0158-4 |
| [8] | Samanta S, Bhushan B, Jain R. Efficiency of naphthalene and salicylate degradation by a recombinant Pseudomonas putida mutant strain defective in glucose metabolism. Applied Microbiology and Biotechnology, 2001, 55(5): 627-631. DOI:10.1007/s002530000553 |
| [9] | Atkinson BW, Mudaly DD, Bux F. Contribution of Pseudomonas spp. to phosphorus uptake in the anoxic zone of an anaerobic-anoxic-aerobic continuous activated sludge system. Water Science and Technology, 2001, 43(1): 139-146. |
| [10] | Gabriel AG, Maurilio LF, Jesús Genaro SM, Angel Isidro CC, Antonio LG. The use of probiotics in aquatic organisms: a review. African Journal of Microbiology Research, 2012, 6(23): 4845-4857. |
| [11] | Guan CR, Cui WJ, Cheng JT, Zhou L, Liu ZM, Zhou ZM. Development of an efficient autoinducible expression system by promoter engineering in Bacillus subtilis. Microbial Cell Factories, 2016, 15: 66. DOI:10.1186/s12934-016-0464-0 |
| [12] | Weir SC, Lee H, Trevors JT. Effect of selected disinfectants on the persistence and movement of a genetically engineered Pseudomonas sp. in soil. Systematic and Applied Microbiology, 1996, 19(3): 421-427. DOI:10.1016/S0723-2020(96)80072-5 |
| [13] | Merroun ML, Raff J, Rossberg A, Hennig C, Reich T, Selenska-Pobell S. Complexation of uranium by cells and S-layer sheets of Bacillus sphaericus JG-A12. Applied and Environmental Microbiology, 2005, 71(9): 5532-5543. DOI:10.1128/AEM.71.9.5532-5543.2005 |
| [14] | Chen DW, Zhang Y, Jiang CY, Liu SJ. Benzoate metabolism intermediate benzoyl coenzyme A affects gentisate pathway regulation in Comamonas testosteroni. Applied and Environmental Microbiology, 2014, 80(13): 4051-4062. DOI:10.1128/AEM.01146-14 |
| [15] | Trögl J, Boušková A, Mrákota J, Pilařová V, Krudencová J, Měchurová J, Kříženecká S, Stloukal R. Removal of nitrates from simulated ion-exchange brines with Paracoccus denitrificans encapsulated in Lentikats biocatalyst. Desalination, 2011, 275(1/3): 82-86. |
| [16] | Zhang H, Li S, Ma B, Huang T, Qiu H, Zhao Z, Huang X, Liu K. Nitrate removal characteristics and 13C metabolic pathways of aerobic denitrifying bacterium Paracoccus denitrificans Z195. Bioresource Technology, 2020, 307: 123230. DOI:10.1016/j.biortech.2020.123230 |
| [17] | Ind AC, Porter SL, Brown MT, Byles ED, De Beyer JA, Godfrey SA, Armitage JP. Inducible-expression plasmid for Rhodobacter sphaeroides and Paracoccus denitrificans. Applied and Environmental Microbiology, 2009, 75(20): 6613-6615. DOI:10.1128/AEM.01587-09 |
| [18] | Inui M, Nakata K, Roh JH, Vertès AA, Yukawa H. Isolation and molecular characterization of pMG160, a mobilizable cryptic plasmid from Rhodobacter blasticus. Applied and Environmental Microbiology, 2003, 69(2): 725-733. DOI:10.1128/AEM.69.2.725-733.2003 |
| [19] | Graupner S, Wackernagel W. A broad-host-range expression vector series including a Ptac test plasmid and its application in the expression of the dod gene of Serratia marcescens (coding for ribulose-5-phosphate 3-epimerase) in Pseudomonas stutzeri. Biomolecular Engineering, 2000, 17(1): 11-16. DOI:10.1016/S1389-0344(00)00061-7 |
| [20] | Andersen JB, Sternberg C, Poulsen LK, Bjrn SP, Molin S. New unstable variants of green fluorescent protein for studies of transient gene expression in bacteria. Applied and Environmental Microbiology, 1998, 64(6): 2240-2246. DOI:10.1128/AEM.64.6.2240-2246.1998 |
| [21] | Kaczmarczyk A, Vorholt JA, Francez-Charlot A. Cumate-inducible gene expression system for sphingomonads and other Alphaproteobacteria. Applied and Environmental Microbiology, 2013, 79(21): 6795-6802. DOI:10.1128/AEM.02296-13 |
| [22] | Tett AJ, Rudder SJ, Bourdès A, Karunakaran R, Poole PS. Regulatable vectors for environmental gene expression in Alphaproteobacteria. Applied and Environmental Microbiology, 2012, 78(19): 7137-7140. DOI:10.1128/AEM.01188-12 |
| [23] | Zhao Y, Lu W, Liu Y, Wang J, Zhou S, Mao Y, Li G, Deng Y. Efficient total nitrogen removal from wastewater by Paracoccus denitrificans DYTN-1. Letters in Applied Microbiology, 2020, 70(4): 263-273. DOI:10.1111/lam.13268 |
| [24] | Hahnke SM, Moosmann P, Erb TJ, Strous M. An improved medium for the anaerobic growth of Paracoccus denitrificans Pd1222. Frontiers in Microbiology, 2014, 5: 18. |
| [25] | Ding NN, Yuan ZQ, Zhang XJ, Chen J, Zhou SH, Deng Y. Programmable cross-ribosome-binding sites to fine-tune the dynamic range of transcription factor-based biosensor. Nucleic Acids Research, 2020, 48(18): 10602-10613. DOI:10.1093/nar/gkaa786 |
| [26] | Ren G, Jin W, Cui K, Rodrigez J, Hu G, Zhang Z, Larson DR, Zhao K. Ctcf-mediated enhancer-promoter interaction is a critical regulator of cell-to-cell variation of gene expression. Molecular Cell, 2017, 67(6): 1049-1058. DOI:10.1016/j.molcel.2017.08.026 |
| [27] | Sun HH, Zhao HM, Ang EL. A new biosensor for stilbenes and a cannabinoid enabled by genome mining of a transcriptional regulator. ACS Synthetic Biology, 2020, 9(4): 698-705. DOI:10.1021/acssynbio.9b00443 |
| [28] | Fernandez-López R, Ruiz R, De La Cruz F, Moncalián G. Transcription factor-based biosensors enlightened by the analyte. Frontiers in Microbiology, 2015, 6: 648. |
| [29] | Giannopoulos G, Sullivan MJ, Hartop KR, Rowley G, Gates AJ, Watmough NJ, Richardson DJ. Tuning the modular Paracoccus denitrificans respirome to adapt from aerobic respiration to anaerobic denitrification. Environmental Microbiology, 2017, 19(12): 4953-4964. DOI:10.1111/1462-2920.13974 |
| [30] | Deng M, Zhao X, Senbati Y, Song K, He X. Nitrogen removal by heterotrophic nitrifying and aerobic denitrifying bacterium Pseudomonas sp. DM02: removal performance, mechanism and immobilized application for real aquaculture wastewater treatment-science direct. Bioresource Technology, 2020, 322: 12455. |
| [31] | Hallin S, Lindgren PE. PCR detection of genes encoding nitrite reductase in denitrifying bacteria. Applied and Environmental Microbiology, 1999, 65(4): 1652-1652. DOI:10.1128/AEM.65.4.1652-1657.1999 |
| [32] | Camargo JA, Alonso A. Ecological and toxicological effects of inorganic nitrogen pollution in aquatic ecosystems: a global assessment. Environment International, 2006, 32(6): 831-849. DOI:10.1016/j.envint.2006.05.002 |
| [33] | Gupta A, Reizman IMB, Reisch CR, Prather KLJ. Dynamic regulation of metabolic flux in engineered bacteria using a pathway-independent quorum-sensing circuit. Nature Biotechnology, 2017, 35(3): 273-279. DOI:10.1038/nbt.3796 |
 2022, Vol. 62
2022, Vol. 62




