中国科学院微生物研究所,中国微生物学会
文章信息
- 韩生义, 卢晓凤, 李淑萍, 林伟山, 胡国元, 石田, 李生庆. 2022
- HAN Shengyi, LU Xiaofeng, LI Shuping, LIN Weishan, HU Guoyuan, SHI Tian, LI Shengqing.
- 产气荚膜梭菌前噬菌体的分布特点及遗传进化分析
- Distribution and genetic evolution of prophages carried by Clostridium perfringens
- 微生物学报, 62(7): 2610-2623
- Acta Microbiologica Sinica, 62(7): 2610-2623
-
文章历史
- 收稿日期:2021-10-22
- 修回日期:2021-12-18
- 网络出版日期:2022-03-24
2. 青海大学农牧学院, 青海 西宁 810003
2. College of Agriculture and Animal Husbandry, Qinghai University, Xining 810003, Qinghai, China
产气荚膜梭菌(Clostridium perfringens)又称魏氏梭菌,是一种产芽胞的革兰氏阳性厌氧菌,对不良环境的抵抗力强,广泛存在于多种环境(土壤、污水、饲料和食物以及人和动物胃肠道等)[1–2]。该菌是一种重要的人畜共患病原菌,可引起人和动物的多种严重疾病,如坏死性肠炎、人畜食物中毒、家畜猝疽、抗生素相关性腹泻及创伤性气性坏疽等,致病性极强且发病快,往往呈急性、来不及表现临床症状就立刻死亡[3–5]。抗生素长期使用和滥用导致产气荚膜梭菌的耐药性问题日益严峻[6],鸡源、猪源和羊源性产气荚膜梭菌的耐药率分别达到66%,82%和100%,且对红霉素、林可霉素、新霉素、四环素和环丙沙星的耐药率很高[7–9],此外耐药基因和毒力基因可通过质粒在产气荚膜梭菌中传播和扩散,使得产气荚膜梭菌的防治更加困难[10–11]。
溶原化是细菌的一种普遍现象,即温和噬菌体感染细菌后,并不裂解杀死细菌,而是将其基因组整合入细菌基因组中,随着细菌的分裂传给后代细菌[12]。溶原性细菌中的前噬菌体影响着宿主的生存方式和对不同环境的适应性(如毒力、耐药性、生物被膜、芽胞的形成等)[13–14],此外,前噬菌体还可以通过诱导进入溶菌周期杀死竞争菌株(即非溶菌菌株),从而提高宿主在种群中的竞争力[15]。对现有的细菌基因组的分析表明,有一半以上细菌基因组存在前噬菌体[16],且不同种属细菌中前噬菌体的分布、基因组特征及遗传进化关系等都不同,并影响细菌的致病性、耐药性和遗传进化等[17–19]。
可移动遗传元件(包括质粒、转座子、整合子、噬菌体和插入序列等)是介导基因水平转移的主要载体和动力,在细菌基因组进化和适应特定环境压力过程中发挥了重要作用[20]。研究表明产气荚膜梭菌基因组携带前噬菌体,并可被诱导产生噬菌体颗粒[21–22]。然而前噬菌体的分布及对基因水平转移的影响仍然是未知的,本研究对产气荚膜梭菌中前噬菌体分布特点、基因组特征及遗传进化关系进行分析,为前噬菌体的多样性及其对产气荚膜梭菌的遗传进化和致病性的影响提供数据和理论依据。
1 材料与方法 1.1 产气荚膜梭菌基因组序列24株产气荚膜梭菌的全基因组数据,均下载自NCBI官网(https://www.ncbi.nlm.nih.gov/genome/browse/#!/prokaryotes/158/),最后访问时间为2021年8月1日。
1.2 前噬菌体预测使用PHASTER(http://phaster.ca/)[23]预测产气荚膜梭菌基因组中前噬菌体序列,将预测结果(包括序列长度、GC含量、完整性、编码蛋白数目、在基因组上的位置及亲缘关系最近的噬菌体等)进行整理,下载前噬菌体序列并重命名(以phicp-Strain-Region的格式进行命名)进行后续分析。
1.3 前噬菌体中ARGs (antibiotics resistance genes)的预测使用抗生素耐药基因数据库(CARD,the comprehensive antibiotic research database, https://card.mcmaster.ca/)[24]和ResFinder 4.1 (https://cge.cbs.dtu.dk/services/ResFinder/)[25]预测产气荚膜梭菌的前噬菌体携带的ARGs。
1.4 前噬菌体中VGs (virulence genes)的预测使用毒力因子数据库(VFDB,virulence factors database, http://www.mgc.ac.cn/VFs/)[26],预测产气荚膜梭菌前噬菌体携带的VGs,预测结果以E-value≤1×10–5确定为目标基因。
1.5 前噬菌体中ABGs/MRGs (antibacterial biocide genes/metal resistance genes)的预测使用抗菌剂和金属抗性基因数据库(BacMet,antibacterial biocide & metal resistance genes database,http://bacmet.bio-medicine.gu.se/)[27],预测产气荚膜梭菌的前噬菌体携带的ABGs/MRGs,预测结果以E-value≤1×10–5确定为目标基因。
1.6 产气荚膜梭菌CRISPR-Cas系统的预测使用细菌CRISPR-Cas系统的在线预测软件CRISPR CasFinder (https://crisprcas.i2bc.paris-saclay.fr/CrisprCasFinder/Index)[28]预测产气荚膜梭菌的CRISPR序列和cas基因,整理和下载预测结果为“确定的”间隔序列。
1.7 前噬菌体的遗传进化关系分析在NCBI上下载已知的26条梭菌噬菌体全基因组序列,并提取主要衣壳蛋白(major capsid protein,MCP)基因和末端酶大亚基(terminase large subunit,TLS)基因序列,使用MEGA 7.0[29]通过邻接法(Neighbor-Joining)构建进化树。
1.8 数据分析前噬菌体之间的ANI (average nucleotide identity)值用OAT软件[30]进行计算,数据用SPSS v19.0[31]进行统计学分析,R version 4.1.1[32]进行作图。
2 结果与分析 2.1 前噬菌体的分布特点从24株产气荚膜梭菌基因组中预测到64条前噬菌体,其中完整的前噬菌体29条,疑似的前噬菌体4条,不完整的前噬菌体31条;前噬菌体长度范围为6.4–65.5 kb,x=28.94 kb,核密度图发现其长度分布有2个峰值,分别为14.5 kb和44.0 kb (图 1A),这2个峰值分别与不完整和完整的前噬菌体序列平均值接近(xincomplete=15.04 kb,xintact=41.20 kb),表明在产气荚膜梭菌基因组中,不完整的前噬菌体序列相对较短,而完整的前噬菌体序列则相对较长;前噬菌体GC%为26.58%–31.45%,x=28.07%,而产气荚膜梭菌基因组GC%为27.95%–31.1%,x=28.50%,两者差异无统计学意义(图 1B);产气荚膜梭菌携带的前噬菌体数量1–5条不等,x=2.67条,多数携带1条或5条前噬菌体(图 1C);前噬菌体占产气荚膜梭菌基因组的0.58%– 5.67%,x=2.23%,66.67%菌株携带的前噬菌体占比不超过2.5% (图 1D)。
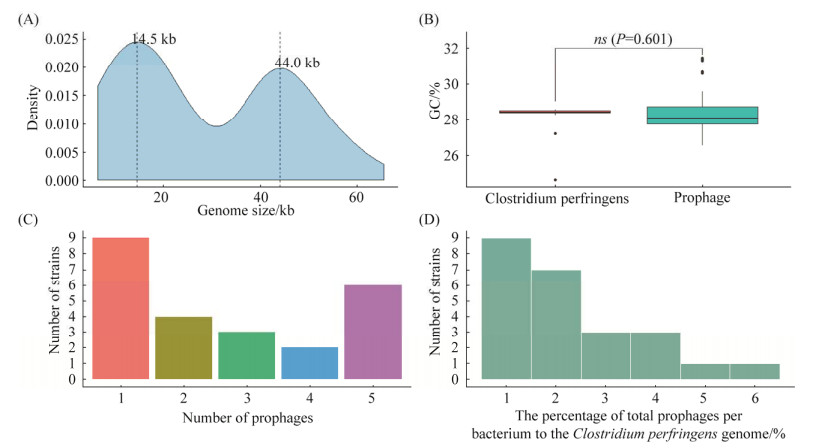
|
| 图 1 前噬菌体的分布特点 Figure 1 Distribution characteristics of prophages. A: genome size of C. perfringens prophages; B: the GC% of C. perfringens and prophages; C: the number of prophages carried in C. perfringens genome; D: the proportional distribution of prophages carried in C. perfringens genome. |
2.2 前噬菌体的分群
前噬菌体ANI值的聚类热图显示,64条前噬菌体聚类为3群(图 2):其中Group 1共23条,群内成员的相似性很高,且完整的前噬菌体占比高(78.26%);Group 2共22条,群内成员的相似性很低,且与另外2群的相似性也很低,仅部分前噬菌体之间的相似性较高,此外该群中不完整的前噬菌体占比很高(占90.9%); Group 3共19条,群内成员的相似性较高,且与Group 1的部分前噬菌体也存在一定的相似性,该群中完整和不完整前噬菌体占比接近(分别为47.74%和52.63%)。

|
| 图 2 前噬菌体聚类热图 Figure 2 The heat map of clustering of prophages. |
2.3 前噬菌体ARGs、VGs和ABGs/MRGs的携带情况
通过分析前噬菌体ARGs、VGs和ABGs/MRGs发现,所有前噬菌体均不携带ARGs;前噬菌体的VGs携带率为17.19% (n=11,携带VGs的完整、疑似和不完整前噬菌体分别为5、1和5条),VGs主要为groEL (编码伴侣蛋白)、nagJ (编码透明质酸酶)、nanH (编码唾液酸酶)、lgtA (编码N-乙酰氨基葡萄糖糖基转移酶)、tufA (编码延伸因子)和tlyC(编码溶血素C),此外,前噬菌体序列phicp-JXJA17-1携带完整的plc基因,该基因编码产气荚膜梭菌的α毒素蛋白,通过BLAST比对发现该基因与已知的产气荚膜梭菌α毒素基因核苷酸序列的查询覆盖度(query coverage)和序列一致性(identity)均达到100%;前噬菌体的ABGs/MRGs的携带率为34.38% (n=22,携带ABGs/MRGs的完整、疑似和不完整前噬菌体分别为9、3和9条),ABGs/MRGs主要为编码铁离子ABC转运蛋白、汞还原酶、烯醇还原酶、AraC家族转录调控因子、多重耐药蛋白、砷酸盐还原酶、锌离子ABC转运蛋白、镁离子和钴离子转运载体等。3个Group中前噬菌体ABGs/MRGs的携带率接近,但Group 3前噬菌体VGs的携带率明显低于Group 1和Group 2 (5.26% vs. 21.74%,22.73%)(图 3)。
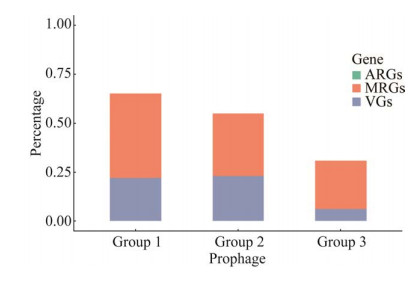
|
| 图 3 不同类群前噬菌体ARGs、VGs和ABGs/MRGs的携带率 Figure 3 The proportional distribution of ARGs, VGs and ABGs/MRGs carried in different groups of prophages. |
2.4 前噬菌体分布与产气荚膜梭菌进化关系及CRISPR-Cas系统之间的关系
从24株产气荚膜梭菌基因组中提取16S rRNA基因序列,用MEGA 7.0构建系统发育进化树,结合产气荚膜梭菌的背景信息分析发现,进化树上红色的类群为A型产气荚膜梭菌(即都能编码α毒素蛋白),毒力和致病性很强;与A型亲缘关系较近的产气荚膜梭菌SM101菌株虽然不编码4种主要毒素蛋白(α、β、ε和ι),但能编码肠毒素,毒力较强;而与A型和SM101亲缘关系较远的产气荚膜梭菌4928STDY7387880既不编码4种主要毒素蛋白,也不编码肠毒素,推测该菌株毒力较弱(图 4A)。Group 2和Group 3前噬菌体在24株产气荚膜梭菌中均有分布,但Group 1前噬菌体仅分布在A型产气荚膜梭菌中,此外,SM101携带的5条前噬菌体均属于Group 2且均为不完整的(1条同时携带透明质酸酶和ABC转运蛋白编码基因,另一条携带ABC转运蛋白编码基因),这5条前噬菌体仅与Group 2内的个别前噬菌体相似性较高,与其他前噬菌体均无相似性;4928STDY7387880携带的5条前噬菌体中4条为不完整的(1条携带溶血素编码基因,另外2条分别携带RNA聚合酶因子和伴侣蛋白dnaK),1条为完整的(同时携带分子伴侣GroEL和ATP酶),除了其中1条与所有前噬菌体均无相似性外,4条与其他前噬菌体均有不同程度的相似性(图 4B和图 2)。24株产气荚膜梭菌的CRISPR-Cas系统分析发现,7株无CRISPR-Cas系统;9株只有1–2个CRISPR序列,且多数CRISPR序列含有3个spacers,仅有一个CRISPR序列含有35个spacers;8株基因组中含有完整的CRISPR-Cas系统,且CRISPR序列中的spacers数量较多,cas基因主要为Type Ⅰ-B (CBA7123、SM101、CP15、FDAARGOS_904和LLY_N11)和Type Ⅱ-C(19TSBNCP、FDAARGOS_904、FORC_003、JXJA17和NCTC13170),除了SM101携带5条前噬菌体外,其他7株含有完整CRISPR-Cas系统的产气荚膜梭菌携带的前噬菌体数量均≤3条。

|
| 图 4 产气荚膜梭菌系统发育树与前噬菌体和CRISPR-Cas之间的关系 Figure 4 The relationship among prophages, CRISPR-Cas system and phylogenetic tree of C. perfringens. A: the phylogenetic tree of 14 C. perfringens strains that based on 16S rRNA gene sequemces, the red branch means C. perfringens type A; B: the groups and population distribution of prophages in C. perfringens; C: the CRISPR-Cas system distribution of C. perfringens. |
2.5 前噬菌体数量分布和CRISPR spacers之间的关系
将产气荚膜梭菌携带的前噬菌体数量与其CRISPR-Cas系统中的spacers数量通过SPSS v19.0进行spearman相关性分析发现,二者之间关系为弱的负相关(图 5),相关系数R=–0.3,说明CRISPR-Cas系统对产气荚膜梭菌携带前噬菌体数量的影响较小。
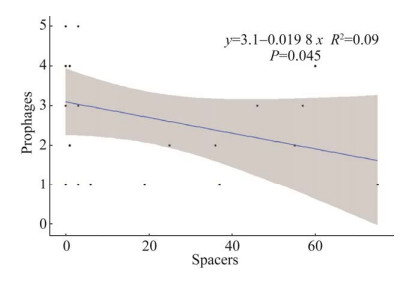
|
| 图 5 产气荚膜梭菌spacers与前噬菌体数量的分布关系 Figure 5 The distribution relationship between spacers and prophages number of C. perfringens. |
2.6 前噬菌体与噬菌体的遗传进化关系
将64株前噬菌体在NCBI进行全基因组BLAST比对后发现,大部分前噬菌体仅与Clostridium phage vB_CpeS-CP51、Clostridium phage phiSM101、Clostridium phage 39-O和Clostridium phage PhiS63基因组存在不同程度的相似性,与其他噬菌体基因组无任何相似性。下载所有已知的26株梭菌噬菌体基因组,其中25株噬菌体的宿主为产气荚膜梭菌,基因组核酸类型均为DNA,长度为17.75–52.65 kb,4株为肌尾科噬菌体,10株为长尾科噬菌体,11株为未分类的有尾目噬菌体;1株噬菌体(Clostridium phage c-st)宿主为肉毒梭菌,基因组核酸类型为DNA,长度为185.68 kb,为长尾科噬菌体。
基于29株完整的前噬菌体与26株已知的梭菌噬菌体构建的全基因组系统进化树结果表明(图 6A),噬菌体和前噬菌体更倾向聚类于各自相对独立的分支;21条前噬菌体与3条噬菌体聚类为一个较大分支(蓝色),7条前噬菌体与1条噬菌体聚类为另一个较小的分支(红色),红色分支中前噬菌体ATCC-2、Del1-5和EHE-NE18-1彼此之间的ANI值较高(83.80%–100.00%),而与蓝色分支中其他Group 1前噬菌体之间ANI值较低(如前噬菌体EHE-NE18-1与其他14条之间ANI值为60.05%–74.98%)或与蓝色分支部分Group 1前噬菌体无任何相似性(如前噬菌体ATCC-2与EHE-NE18-3之间ANI值为0,前噬菌体Del1-5与Del1-1和JP55之间ANI值为0),此外前噬菌体CBA7123、FJP838-5和4928STDY7387880-5仅与ATCC-2、Del1-5、EHE-NE18-1和FORC_003-2之间的ANI值较高,使得Group 1中的这7条前噬菌体更倾向于聚类到红支;红蓝分支中的前噬菌体不仅与多数噬菌体的遗传进化距离较远,而且两支彼此之间的遗传进化距离也较远;蓝色分支中的7条前噬菌体仅与Clostridium phage vB_CpeS-CP51亲缘关系非常近,位于一个小的分支,Clostridium phage vB_CpeS-CP51是一株长尾科的溶原性噬菌体,宿主为Clostridium perfringens 5147-97,基因组为双链DNA,长度为39 108 bp,GC%为28.08%,编码50个蛋白;红色分支中的7条前噬菌体与Clostridium phage phiSM101亲缘关系很近,Clostridium phage phiSM101也是一株长尾科的噬菌体,宿主为Clostridium perfringens SM101,基因组为双链DNA,长度为38 092 bp,GC%为28.13%,编码54个蛋白。基于前噬菌体和噬菌体主要衣壳蛋白(MCPs)与末端酶大亚基(TCLs)基因序列构建的系统进化树也表明,前噬菌体的MCPs和TCLs也都更倾向于聚类在同一分支(图 6B黄色分支,图 6C绿色与蓝色分支),即前噬菌体彼此之间不论是结构蛋白还是主要的酶类的遗传进化关系都很近,而与多数噬菌体的遗传进化关系较远。

|
图 6 完整前噬菌体和噬菌体的系统发育进化树 Figure 6 The phylogenetic tree of intact prophages and phages. A: the phylogenetic tree based on genome; B: the phylogenetic tree based on major capsid protein; C: the phylogenetic tree based on terminase large subunit.      |
2.7 前噬菌体与产气荚膜梭菌噬菌体之间的基因组比较分析
以Clostridium phage vB_CpeS-CP51为参考基因组,选取与其位于同一分支的7条前噬菌体序列进行全基因组比对,结果表明7条前噬菌体与Clostridium phage vB_CpeS-CP51的部分结构蛋白(portal protein、minor capsid protein、major capsid protein、capsid protein、head-tail connector protein、major tail protein、tape measure protein和phage tail protein)、酶类(terminase large subunit和holin)以及一些功能未知的假定蛋白的编码基因的同源性较高,其他基因的同源性很低(图 7)。
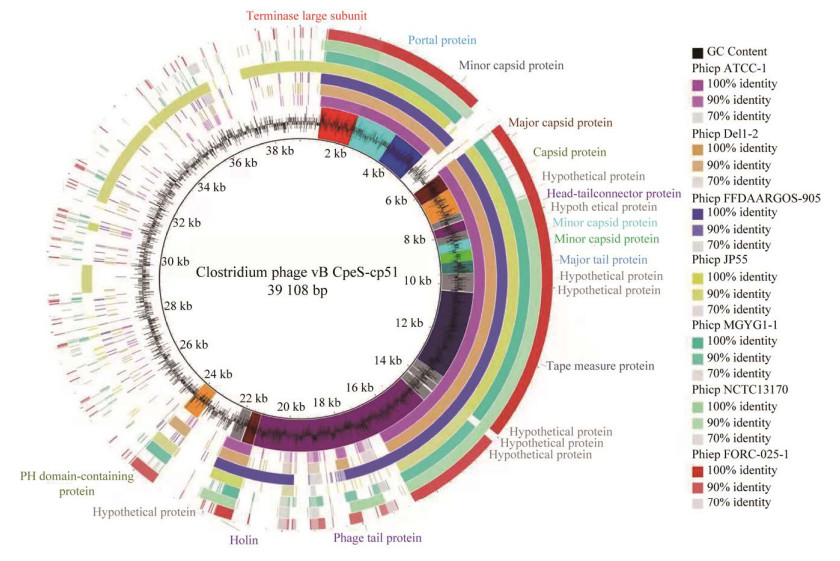
|
| 图 7 噬菌体基因组和7条前噬菌体的全基因组比对 Figure 7 The genome comparison between the phage and 7 prophages, and the Clostridium phage VB_cpES-CP51 as the reference genome. |
3 讨论
溶原化是噬菌体与细菌之间的军备竞赛过程中的一种独特的作用结果,很多细菌是非溶原性细菌,而溶原性细菌携带的前噬菌体除了在细菌之间传播毒力基因和耐药基因外,还可以促进溶原性细菌某些基因的表达[33–34]。研究发现,溶原菌的占比与细菌的种属有关,如大肠杆菌中溶原菌占比为100%[35],肺炎链球菌中溶原菌占比为100%[36],鲍曼不动杆菌中溶原菌占比为99.5%[37],而变异链球菌中溶原菌占比则为13.45%[38],产气荚膜梭菌中溶原菌的占比也很高(100%),但其平均携带的前噬菌体数量较少(2.67条),而大肠杆菌、沙门氏菌和鲍曼不动杆菌的平均携带的前噬菌体分别为8.2、5.6和4.0条[36, 39];前噬菌体在许多细菌中占比很高,最多可占宿主基因组的10%–20%[40],本研究中前噬菌体占产气荚膜梭菌基因组比例为2.23%,明显低于埃希氏菌属、链球菌属、螺旋体属(10%–20%)和沙门氏菌属(4.9%)[40–41],低于洋葱伯克霍尔德氏菌(3.67%),接近于假伯克霍尔德氏菌(2.34%)[42]。产气荚膜梭菌的前噬菌体长度分布的2个峰值与完整和不完整前噬菌体的平均长度接近,而所有前噬菌体的GC%与产气荚膜梭菌的GC%接近,这与之前的研究结果一致,即由于噬菌体完全依靠宿主的翻译机制进行转录和翻译,因此噬菌体更倾向具有和宿主接近的GC%和密码子使用模式[43],而双峰分布模式同样证实了噬菌体基因组整合到宿主基因组后,由于完整的前噬菌体会在某些条件下被激活进入裂解周期杀死宿主菌,在长期进化过程中,完整的前噬菌体会被宿主菌删除某些关键基因(如部分结构基因等)变成不完整而被灭活,宿主菌通过这种驯化机制解除了被裂解杀死的风险,同时保留了前噬菌体中的某些对自身有益的基因(如耐药基因和毒力基因等)[39]。
研究表明,噬菌体能携带ARGs并通过普遍性转导介导ARGs在细菌间转移,如在鲍曼不动杆菌中,前噬菌体携带了9个不同家族的ARGs (如β-内酰胺类、四环素类和萘啶酸类等)[44],产肠毒素大肠杆菌中的前噬菌体携带β-内酰胺酶耐药基因(blaTEM-1B),并通过普遍性转导介导了ARGs的转移[35],我们发现产气荚膜梭菌的前噬菌体不携带任何ARGs,推测在产气荚膜梭菌中ARGs可能通过质粒、转座子或整合子等其他可移动遗传元件进行转移。目前研究发现,产气荚膜梭菌至少有23种毒素和毒力相关酶类,其中α、β、ε和ι为主要致死毒素,而α毒素基因cpa/plc位于染色体上,是毒性最强、最重要的毒素,其他毒素基因都位于质粒上[11, 45];很多菌株还会产生产气荚膜梭菌肠毒素(C. perfringens enterotoxin,CPE)、β2毒素、溶血素(PFO)、坏死性肠炎毒素(NetB)和大分子细胞毒素(TpeL)等[46]。本研究发现,前噬菌体除了会携带α毒素基因,还会携带其他毒力基因,这些毒力因子除了直接赋予产气荚膜梭菌强致死性外,还会通过增强在感染动物体内的黏附、定殖及扩散能力,进一步增强产气荚膜梭菌的致病性,如唾液酸酶能帮助产气荚膜梭菌黏附到小肠上皮细胞并能增强ε毒素的毒性[47]。噬菌体是细菌在污染环境中重金属抗性基因的重要储存库,其中77.0%的膜转运蛋白基因和61.7%的金属还原酶基因都位于溶原性噬菌体上,携带ABGs/MRGs的噬菌体通过整合到细菌基因组,提高了细菌对重金属的抗性[48]。本研究发现,前噬菌体携带了多种金属离子还原酶和转运蛋白,推测前噬菌体对产气荚膜梭菌在适应不良环境和化学胁迫因素方面也具有重要的影响。
根据毒素(α、β、ε和ι为)的产生情况,产气荚膜梭菌被分为A (α)型、B (α、β、ε)型、C (α、β)型、D (α、ε)型和E (α、ι)型,其中A型产气荚膜梭菌主要引起人和动物气性坏疽和食物中毒,还可引起牛、羔羊、山羊、驯鹿、仔猪和家兔等的肠毒血症[5]。本研究发现前噬菌体的分布与产气荚膜梭菌的遗传进化有密切关系,其中Group 1前噬菌体仅存在于A型产气荚膜梭菌中且多为完整的,Group 2中前噬菌体多为不完整的,Group 3中前噬菌体完整和不完整的各占近半;由于细菌在进化过程中会删除前噬菌体的某些基因,使其成为不完整的前噬菌体[35],推测在产气荚膜梭菌的进化过程中,Group 2前噬菌体是最早出现并感染产气荚膜梭菌,Group 3次之,Group 1前噬菌体较晚出现且宿主谱相对较窄,只能感染A型产气荚膜梭菌。除了SM101,其他具有完整CRISPR-Cas系统的产气荚膜梭菌携带的前噬菌体数量较少(1–3)条,前噬菌体数量和spacers之间的关系为弱的负相关,这与之前的研究结果一致[15],说明CRISPR-Cas系统对前噬菌体的数量影响作用较小,其他防御系统(如R-M系统、流产感染系统、BREX系统、毒素-抗毒素系统等)在抗噬菌体感染过程中发挥了主要作用[49]。不论基于前噬菌体全基因组还是特定基因(主要衣壳蛋白和末端酶大亚基)构建的系统进化树结果都表明,产气荚膜梭菌的前噬菌体与已知的产气荚膜梭菌的噬菌体的遗传进化距离和亲缘关系较远,即便与亲缘关系最近的噬菌体也仅有部分基因存在较高的同源性,这说明产气荚膜梭菌的前噬菌体是一类新的噬菌体,其功能和对宿主遗传进化影响仍需要进行深入研究。
4 结论产气荚膜梭菌普遍携带前噬菌体,前噬菌体的分布特点与产气荚膜梭菌的遗传进化关系存在联系,但CRISPR-Cas系统对前噬菌体的数量影响较小,前噬菌体不携带耐药基因,但其携带的毒力基因和重金属代谢相关等基因对产气荚膜梭菌的毒力因子获得和转移、致病性演变及环境适应能力具有重要影响。
| [1] | Martel A, Devriese LA, Cauwerts K, de Gussem K, Decostere A, Haesebrouck F. Susceptibility of Clostridium perfringens strains from broiler chickens to antibiotics and anticoccidials. Avian Pathology, 2004, 33(1): 3-7. DOI:10.1080/0307945031000163291 |
| [2] | Abbona CC, Stagnitta PV. Clostridium perfringens: comparative effects of heat and osmotic stress on non-enterotoxigenic and enterotoxigenic strains. Anaerobe, 2016, 39: 105-113. DOI:10.1016/j.anaerobe.2016.03.007 |
| [3] | Lebrun M, Mainil JG, Linden A. Cattle enterotoxaemia and Clostridium perfringens: description, diagnosis and prophylaxis. Veterinary Record, 2010, 167(1): 13-22. DOI:10.1136/vr.167.1.12 |
| [4] | Guran HS, Oksuztepe G. Detection and typing of Clostridium perfringens from retail chicken meat parts. Letters in Applied Microbiology, 2013, 57(1): 77-82. DOI:10.1111/lam.12088 |
| [5] | Stevens DL, Aldape MJ, Bryant AE. Life-threatening clostridial infections. Anaerobe, 2012, 18(2): 254-259. DOI:10.1016/j.anaerobe.2011.11.001 |
| [6] | Watkins KL, Shryock TR, Dearth RN, Saif YM. In-vitro antimicrobial susceptibility of Clostridium perfringens from commercial Turkey and broiler chicken origin. Veterinary Microbiology, 1997, 54(2): 195-200. DOI:10.1016/S0378-1135(96)01276-X |
| [7] | Gholamiandehkordi A, Eeckhaut V, Lanckriet A, Timbermont L, Bjerrum L, Ducatelle R, Haesebrouck F, van Immerseel F. Antimicrobial resistance in Clostridium perfringens isolates from broilers in Belgium. Veterinary Research Communications, 2009, 33(8): 1031-1037. DOI:10.1007/s11259-009-9306-4 |
| [8] | Ngamwongsatit B, Tanomsridachchai W, Suthienkul O, Urairong S, Navasakuljinda W, Janvilisri T. Multidrug resistance in Clostridium perfringens isolated from diarrheal neonatal piglets in Thailand. Anaerobe, 2016, 38: 88-93. DOI:10.1016/j.anaerobe.2015.12.012 |
| [9] | Mohiuddin M, Iqbal Z, Siddique A, Liao SQ, Salamat MKF, Qi NS, Din AM, Sun MF. Prevalence, genotypic and phenotypic characterization and antibiotic resistance profile of Clostridium perfringens type A and D isolated from feces of sheep (Ovis aries) and goats (Capra hircus) in punjab, Pakistan. Toxins, 2020, 12(10): 657. DOI:10.3390/toxins12100657 |
| [10] | Adams V, Han XY, Lyras D, Rood JI. Antibiotic resistance plasmids and mobile genetic elements of Clostridium perfringens. Plasmid, 2018, 99: 32-39. DOI:10.1016/j.plasmid.2018.07.002 |
| [11] | Li JH, Adams V, Bannam TL, Miyamoto K, Garcia JP, Uzal FA, Rood JI, McClane BA. Toxin plasmids of Clostridium perfringens. Microbiology and Molecular Biology Reviews: MMBR, 2013, 77(2): 208-233. DOI:10.1128/MMBR.00062-12 |
| [12] | Brüssow H, Canchaya C, Hardt WD. Phages and the evolution of bacterial pathogens: from genomic rearrangements to lysogenic conversion. Microbiology and Molecular Biology Reviews: MMBR, 2004, 68(3): 560-602. DOI:10.1128/MMBR.68.3.560-602.2004 |
| [13] | Fortier LC, Sekulovic O. Importance of prophages to evolution and virulence of bacterial pathogens. Virulence, 2013, 4(5): 354-365. DOI:10.4161/viru.24498 |
| [14] | Harrison E, Brockhurst MA. Ecological and evolutionary benefits of temperate phage: what does or doesn't kill you makes you stronger. BioEssays, 2017, 39(12): 1700112. DOI:10.1002/bies.201700112 |
| [15] | Paul JH. Prophages in marine bacteria: dangerous molecular time bombs or the key to survival in the seas?. The ISME Journal, 2008, 2(6): 579-589. DOI:10.1038/ismej.2008.35 |
| [16] | Canchaya C, Proux C, Fournous G, Bruttin A, Brüssow H. Prophage genomics. Microbiology and Molecular Biology Reviews, 2003, 67(2): 238-276. DOI:10.1128/MMBR.67.2.238-276.2003 |
| [17] | Eraclio G, Fortina MG, Labrie SJ, Tremblay DM, Moineau S. Characterization of prophages of Lactococcus garvieae. Scientific Reports, 2017, 7: 1856. DOI:10.1038/s41598-017-02038-y |
| [18] | Diene SM, Corvaglia AR, François P, van der Mee-Marquet N, Regional Infection Control Group of the Centre Region. Prophages and adaptation of Staphylococcus aureus ST398 to the human clinic. BMC Genomics, 2017, 18(1): 133. DOI:10.1186/s12864-017-3516-x |
| [19] | Casjens SR, Grose JH. Contributions of P2- and P22-like prophages to understanding the enormous diversity and abundance of tailed bacteriophages. Virology, 2016, 496: 255-276. DOI:10.1016/j.virol.2016.05.022 |
| [20] | Frost LS, Leplae R, Summers AO, Toussaint A. Mobile genetic elements: the agents of open source evolution. Nature Reviews Microbiology, 2005, 3(9): 722-732. DOI:10.1038/nrmicro1235 |
| [21] | Zimmer M, Vukov N, Scherer S, Loessner MJ. The murein hydrolase of the bacteriophage phi3626 dual lysis system is active against all tested Clostridium perfringens strains. Applied and Environmental Microbiology, 2002, 68(11): 5311-5317. DOI:10.1128/AEM.68.11.5311-5317.2002 |
| [22] | Gervasi T, Curto RL, Narbad A, Mayer MJ. Complete genome sequence of ΦCP51, a temperate bacteriophage of Clostridium perfringens. Archives of Virology, 2013, 158(9): 2015-2017. DOI:10.1007/s00705-013-1647-1 |
| [23] | Arndt D, Grant JR, Marcu A, Sajed T, Pon A, Liang YJ, Wishart DS. PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Research, 2016, 44(W1): W16-W21. DOI:10.1093/nar/gkw387 |
| [24] | Alcock BP, Raphenya AR, Lau TTY, Tsang KK, Bouchard M, Edalatmand A, Huynh W, Nguyen ALV, Cheng AA, Liu SH, Min SY, Miroshnichenko A, Tran HK, Werfalli RE, Nasir JA, Oloni M, Speicher DJ, Florescu A, Singh B, Faltyn M, Hernandez-Koutoucheva A, Sharma AN, Bordeleau E, Pawlowski AC, Zubyk HL, Dooley D, Griffiths E, Maguire F, Winsor GL, Beiko RG, Brinkman FSL, Hsiao WWL, Domselaar GV, McArthur AG. CARD 2020: antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Research, 2020, 48(1): D517-D525. |
| [25] | Bortolaia V, Kaas RS, Ruppe E, Roberts MC, Schwarz S, Cattoir V, Philippon A, Allesoe RL, Rebelo AR, Florensa AF, Fagelhauer L, Chakraborty T, Neumann B, Werner G, Bender JK, Stingl K, Nguyen M, Coppens J, Xavier BB, Malhotra-Kumar S, Westh H, Pinholt M, Anjum MF, Duggett NA, Kempf I, Nykäsenoja S, Olkkola S, Wieczorek K, Amaro A, Clemente L, Mossong J, Losch S, Ragimbeau C, Lund O, Aarestrup FM. ResFinder 4.0 for predictions of phenotypes from genotypes. The Journal of Antimicrobial Chemotherapy, 2020, 75(12): 3491-3500. |
| [26] | Chen LH, Zheng DD, Liu B, Yang J, Jin Q. VFDB 2016: hierarchical and refined dataset for big data analysis—10 years on. Nucleic Acids Research, 2015, 44(D1): D694-D697. |
| [27] | Pal C, Bengtsson-Palme J, Rensing C, Kristiansson E, Larsson DGJ. BacMet: antibacterial biocide and metal resistance genes database. Nucleic Acids Research, 2013, 42(D1): D737-D743. |
| [28] | Couvin D, Bernheim A, Toffano-Nioche C, Touchon M, Michalik J, Néron B, Rocha EPC, Vergnaud G, Gautheret D, Pourcel C. CRISPRCasFinder, an update of CRISRFinder, includes a portable version, enhanced performance and integrates search for Cas proteins. Nucleic Acids Research, 2018, 46(W1): W246-W251. DOI:10.1093/nar/gky425 |
| [29] | Kumar S, Stecher G, Tamura K. MEGA 7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Molecular Biology and Evolution, 2016, 33(7): 1870-1874. DOI:10.1093/molbev/msw054 |
| [30] | Lee I, Ouk Kim Y, Park SC, Chun J. OrthoANI: an improved algorithm and software for calculating average nucleotide identity. International Journal of Systematic and Evolutionary Microbiology, 2016, 66(2): 1100-1103. DOI:10.1099/ijsem.0.000760 |
| [31] | Liu RX, Kuang J, Gong Q, Hou XL. Principal component regression analysis with spss. Computer Methods and Programs in Biomedicine, 2003, 71(2): 141-147. DOI:10.1016/S0169-2607(02)00058-5 |
| [32] | Tippmann S. Programming tools: adventures with R. Nature, 2015, 517(7532): 109-110. DOI:10.1038/517109a |
| [33] | Touchon M, Bernheim A, Rocha EP. Genetic and life-history traits associated with the distribution of prophages in bacteria. The ISME Journal, 2016, 10(11): 2744-2754. DOI:10.1038/ismej.2016.47 |
| [34] | Feiner R, Argov T, Rabinovich L, Sigal N, Borovok I, Herskovits AA. A new perspective on lysogeny: prophages as active regulatory switches of bacteria. Nature Reviews Microbiology, 2015, 13(10): 641-650. DOI:10.1038/nrmicro3527 |
| [35] | Wang MZ, Zeng ZL, Jiang FW, Zheng Y, Shen HG, Macedo N, Sun YX, Sahin O, Li GW. Role of enterotoxigenic Escherichia coli prophage in spreading antibiotic resistance in a porcine-derived environment. Environmental Microbiology, 2020, 22(12): 4974-4984. DOI:10.1111/1462-2920.15084 |
| [36] | Brueggemann AB, Harrold CL, Rezaei Javan R, van Tonder AJ, McDonnell AJ, Edwards BA. Pneumococcal prophages are diverse, but not without structure or history. Scientific Reports, 2017, 7: 42976. DOI:10.1038/srep42976 |
| [37] | Costa AR, Monteiro R, Azeredo J. Genomic analysis of Acinetobacter baumannii prophages reveals remarkable diversity and suggests profound impact on bacterial virulence and fitness. Scientific Reports, 2018, 8: 15346. DOI:10.1038/s41598-018-33800-5 |
| [38] | Fu TW, Fan XY, Long QX, Deng WY, Song JL, Huang EY. Comparative analysis of prophages in Streptococcus mutans genomes. PeerJ, 2017, 5: e4057. DOI:10.7717/peerj.4057 |
| [39] | Bobay LM, Touchon M, Rocha EPC. Pervasive domestication of defective prophages by bacteria. Proceedings of the National Academy of Sciences of the United States of America, 2014, 111(33): 12127-12132. DOI:10.1073/pnas.1405336111 |
| [40] | Casjens S. Prophages and bacterial genomics: what have we learned so far?. Molecular Microbiology, 2003, 49(2): 277-300. DOI:10.1046/j.1365-2958.2003.03580.x |
| [41] | Bobay LM, Rocha EPC, Touchon M. The adaptation of temperate bacteriophages to their host genomes. Molecular Biology and Evolution, 2012, 30(4): 737-751. |
| [42] | Weiser R, Yap ZL, Otter A, Jones BV, Salvage J, Parkhill J, Mahenthiralingam E. A novel inducible prophage from Burkholderia vietnamiensis G4 is widely distributed across the species and has lytic activity against pathogenic Burkholderia. Viruses, 2020, 12(6): 601. DOI:10.3390/v12060601 |
| [43] | Chithambaram S, Prabhakaran R, Xia XH. Differential codon adaptation between dsDNA and ssDNA phages in Escherichia coli. Molecular Biology and Evolution, 2014, 31(6): 1606-1617. DOI:10.1093/molbev/msu087 |
| [44] | López-Leal G, Santamaria RI, Cevallos MÁ, Gonzalez V, Castillo-Ramírez S. Letter to the editor: prophages encode antibiotic resistance genes in Acinetobacter baumannii. Microbial Drug Resistance, 2020, 26(10): 1275-1277. DOI:10.1089/mdr.2019.0362 |
| [45] | Revitt-Mills SA, Rood JI, Adams V. Clostridium perfringens extracellular toxins and enzymes: 20 and counting. Microbiology Australia, 2015, 36(3): 114. DOI:10.1071/MA15039 |
| [46] | Kiu R, Hall LJ. An update on the human and animal enteric pathogen Clostridium perfringens. Emerging Microbes & Infections, 2018, 7(1): 1-15. |
| [47] | Li JH, McClane BA. The Sialidases of Clostridium perfringens type D strain CN3718 differ in their properties and sensitivities to inhibitors. Applied and Environmental Microbiology, 2014, 80(5): 1701-1709. DOI:10.1128/AEM.03440-13 |
| [48] | Huang D, Yu PF, Ye M, Schwarz C, Jiang X, Alvarez PJJ. Enhanced mutualistic symbiosis between soil phages and bacteria with elevated chromium-induced environmental stress. Microbiome, 2021, 9(1): 150. DOI:10.1186/s40168-021-01074-1 |
| [49] | Koonin EV, Makarova KS, Wolf YI. Evolutionary genomics of defense systems in archaea and bacteria. Annual Review of Microbiology, 2017, 71: 233-261. DOI:10.1146/annurev-micro-090816-093830 |
 2022, Vol. 62
2022, Vol. 62




