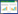中国科学院微生物研究所,中国微生物学会
文章信息
- 徐旸, 王锦鸿, 徐桐, 向华, 韩静. 2023
- XU Yang, WANG Jinhong, XU Tong, XIANG Hua, HAN Jing.
- 嗜盐古菌启动子的研究及应用
- Research and application of haloarchaeal promoter
- 微生物学报, 63(2): 523-539
- Acta Microbiologica Sinica, 63(2): 523-539
-
文章历史
- 收稿日期:2022-06-06
- 网络出版日期:2022-08-17
2. 中国科学院大学生命科学学院, 北京 100049
2. College of Life Sciences, University of Chinese Academy of Sciences, Beijing 100049, China
古菌(archaea)是与细菌和真核生物并列的3种生命形式之一,是一类主要在极端环境(高温、高盐、高酸和高压等)中生活的单细胞原核生物[1]。基于其具有三域生命的融合特征及其自身的独特性、对极端环境的适应性和生理代谢的特殊性等特点,古菌在生命进化、地球生态系统组成、前沿生物技术开发以及合成生物学领域都具有重要的研究价值[2-4]。嗜盐古菌(halophilic archaea)是古菌的一个代表性生理类群,生活在地球上的各种高盐环境中,相比其它古菌类群,拥有易培养、不易被杂菌污染、遗传与代谢类型丰富、遗传操作系统完善等优势,被作为研究古菌环境适应和遗传代谢等方面的理想材料[5]。此外,嗜盐古菌普遍具有合成生物可降解塑料聚羟基脂肪酸酯(polyhydroxyalkanoates, PHA)的能力,且具有可通过水裂解实现胞内产物提取等特点,因此它们在PHA生产中显示出良好的应用前景[6]。在现代分子遗传学研究中,遗传操作元件及工具的开发为上述研究提供了必要的技术手段。其中,作为一种重要遗传元件的嗜盐古菌启动子,在过去30年已经进行了一定程度的研究和开发。
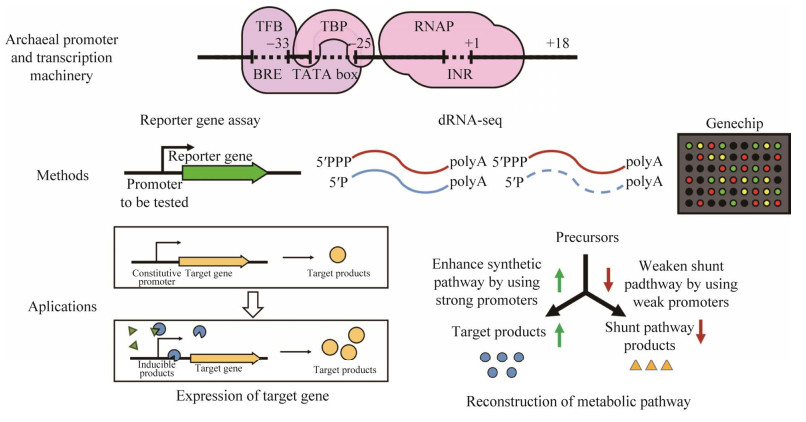
本文从古菌启动子的结构和功能出发,总结和介绍了古菌的转录机器和转录起始过程、嗜盐古菌启动子研究中的常用方法、列举了几种研究较为充分的组成型和诱导型嗜盐古菌启动子及其应用实例(图 1),并对嗜盐古菌启动子的开发应用价值和进一步的研究方向进行了展望。
|
| 图 1 (嗜盐)古菌启动子及其基本转录机器和应用 Figure 1 Structure and function of archaeal promoter in transcription and applications of haloarchaeal promoter. |
1 古菌的启动子与转录 1.1 古菌启动子
启动子(promoter)是基因表达调控的重要遗传元件,其本质为RNA聚合酶识别、结合和开始转录的一段基因序列。如表 1所示,细菌的启动子核心元件包括–10区(TATA盒)和–35区,真核细胞的启动子核心元件包括TATA盒(TATA box)[7]、转录因子IIB (transcription factor IIB, TFIIB)识别元件(TFIIB recognition element, BRE)[8]以及起始子元件(initiator, INR)[9]。细菌启动子与真核生物启动子往往因结构特征不同而不兼容。
| Features | Bacteria | Archaea | Eukarya |
| Core promoter elements | –10 and –35 elements | TATA box, BRE, and INR | TATA box, BRE, and INR |
| RNA polymerase | One type, low subunit complexity | One type, highly homologous with RNAP II, high subunit complexity | Three types (RNAP I, II, and III), high subunit complexity |
| General transcriptional factors | with or without σ factor | TBP, TFB, and TFE | TFIIB, TFIID, TFIIE, TFIIF, and TFIIH (RNAP II) |
与真核细胞启动子相似,古菌的启动子结构包括TATA盒、转录因子B (transcription factor B, TFB)识别元件(BRE)和起始子元件(INR) (图 1, 表 1)。TATA盒富含AT碱基,一般位于基因转录起始位点上游25–30 bp处,转录起始时可被TATA盒结合蛋白(TATA-binding protein, TBP)识别并结合[10-13]。TATA盒对编码rRNA、tRNA以及编码蛋白基因的转录起始都非常重要,它也是决定启动子活性和强度的关键因素[11]。BRE富含嘌呤,通常位于TATA盒上游,它介导了TFB与启动子序列的特异性结合,对于转录前起始复合物的组装和转录起始方向的确定起到重要作用[13-15]。
1.2 古菌的转录机器与转录起始过程如表 1所示,古菌的转录机器与真核生物更为相似,它的基本转录装置包括启动子(见1.2)、DNA依赖的RNA聚合酶(RNA polymerase, RNAP)和基础转录因子(general transcription factor, GTF)。古菌的RNAP通常包含10–13个亚基,绝大部分亚基序列与真核生物RNAP亚基高度同源[16]。最常见的古菌GTF包括TBP与TFB,其次为转录因子E (transcription factor E, TFE),它们分别与真核生物的TBP、TFIIB以及TFIIEα氨基端的部分序列同源[17-19]。
古菌转录起始时,首先由TBP识别并结合TATA盒。然后TFB的羧基末端结合TATA盒上游的BRE序列,氨基端与TATA盒下游序列接触,以此确定转录方向。TFB与BRE相互作用,招募RNAP,形成TBP-TFB-RNAP转录前起始复合物。随后,转录在线性DNA模板上开始。其它转录因子(TFA、TFE、TFF和TFH等)对招募RNAP及DNA的解链具有促进作用,如TFE促进DNA的解旋和RNAP的构象改变[20],形成开放复合物,使转录起始。与细菌转录起始过程相似,古菌在形成开放复合物的过程中无需其它的转录因子或ATP水解提供能量,而在真核生物的这一过程中,转录因子TFIIH的DNA解旋酶活性需要ATP水解供能[21-22]。
综上所述,古菌启动子结构、基础转录机器和转录起始过程与真核细胞高度相似且更简单。但是,古菌通常采用与细菌类似的策略实现基因的转录表达调控[23]。因此,古菌启动子及古菌基因转录调控研究,不仅为揭示古菌转录调控机制提供重要的理论依据,而且对理解包含启动子在内的基因调控序列进化乃至生物系统进化具有重要的研究价值。
2 启动子研究的常用方法及其在嗜盐古菌启动子研究中的应用在启动子的研究中,常用的技术方法主要包括报告基因法、DNA微阵列法和RNA-seq (图 1),它们被用于启动子强度测试、启动子核心元件鉴定、启动子筛选等过程。
2.1 报告基因法报告基因法是一种研究基因转录活性和表达水平的常用方法,具有快速、直观、灵敏和稳定的特点。原理为将报告基因置于受试启动子后,通过测定其表达强度即可定性或定量表征启动子的强度。在嗜盐古菌中,常用的报告基因包含bgaH、pyrE2、dhfr与gfp四种基因。
bgaH基因由Holmes等从阿利坎特富盐菌(Haloferax alicantei, Hfx. alicantei)的突变体中分离出来[24],该基因编码β-半乳糖苷酶[25],可将无色化合物5-溴-4-氯-3-吲哚-β-d-吡喃半乳糖苷(X-gal)切割为半乳糖和深蓝色物质5-溴-4-靛蓝。陈向东团队利用了bgaH作为报告基因鉴定了PamyH在沃氏富盐菌(Haloferax volcanii, Hfx. volcanii)中的转录活性[26]。沈萍团队从盐生盐杆菌(Halobacterium halobium, Hbt. halobium)中发现了同时具有细菌和古菌启动子融合特征的启动子,并利用bgaH报告基因发现它们在大肠杆菌(Escherichia coli, E. coli)和酿酒酵母(Saccharomyces cerevisiae, S. cerevisiae)中都具有启动子功能[27]。嗜盐古菌有尾双链DNA病毒HF2的基因组上存在9个基因间重复序列(intergenic repeat, IR)[28-29]。近期,Dyall-Smith等利用bgaH报告系统测试发现它们均具有启动子活性[30-31]。
pyrE基因编码乳清酸磷酸核糖转移酶(orotate phosphoribosyltransferase, OPRT),催化乳清酸生成5-磷酸-乳清酸核苷酸,参与尿嘧啶的生物合成。Hfx. volcanii的突变体ΔpyrE2是尿嘧啶营养缺陷型菌株,将受试启动子启动pyrE2转录的质粒转入ΔpyrE2中,在不含尿嘧啶的培养基中培养,通过菌株的生长来表征启动子的活性[32]。Lund团队在研究来自Hfx. volcanii的色氨酸诱导型启动子Ptna的活性时,同时使用了bgaH和pyrE2两种相互独立的报告基因系统定性和定量研究了该启动子的活性,验证了Ptna具有较高的诱导活性[32]。同时,他们还利用pyrE2报告基因测试了Ptna在Hfx. volcanii中对色氨酸诱导的严谨性和高效性[33]。
dhfr基因编码二氢叶酸还原酶(dihydrofolate reductase, DHFR),在胞内将二氢叶酸还原为具有生物活性的四氢叶酸[34]。甲氧苄啶(trimethoprim, TMP)是DHFR的竞争性抑制剂,抑制菌体的生长。在细胞中过表达DHFR时,菌株对TMP的耐受性提高,且耐受程度与胞内DHFR的表达量呈正相关[35-36]。Danner和Soppa利用来自Hfx. volcanii的dhfr作为报告基因,通过对比Hfx. volcanii对TMP的耐受程度,鉴定了盐场盐杆菌(Halobacterium salinarium, Hbt. salinarium)的铁氧还蛋白基因启动子(Pfdx)的核心元件TATA盒[37]。
gfp基因编码绿色荧光蛋白(green fluorescent protein, GFP),来自维多利亚多管发光水母(Aequorea victoria, A. victoria)的gfp是最常用的报告基因[38],具有观察或检测时无需添加底物,可用于亚细胞定位的优点[39]。GFP蛋白的荧光强度与GFP的表达量成正比,方便定量表征启动子强度。嗜盐古菌中使用突变体蛋白可溶性红移绿色荧光蛋白(soluble modified red-shifted GFP, smRS-GFP)作为荧光蛋白,突变蛋白在细胞质中的溶解度增大,吸收波长也进行了红移[40-42]。本文作者所在团队利用smRS-GFP作为报告系统,通过定量GFP荧光强度成功鉴定到地中海富盐菌(Haloferaxmediterranei, Hfx. mediterranei)的PphaR启动子的顺式作用元件[43]。
2.2 DNA微阵列法DNA微阵列也被称为基因芯片技术,它可在基因组规模上获取特定条件下的基因表达数据[44-45]。Lund团队在Hfx. volcanii中通过该技术筛选出了受色氨酸诱导的启动子Ptna[33],目前该启动子在嗜盐古菌研究中得到了广泛应用[46]。本文作者所在团队在研究Hfx. mediterranei中参与果糖转运的磷酸烯醇式丙酮酸依赖的磷酸转移酶系统(phosphoenolpyruvate phosphotransferase system, PTS)时,利用DNA微阵列分析方法检测了果糖诱导时的基因差异表达,证明了调控蛋白GlpR是PTS系统重要的调控因子[47]。
2.3 RNA-seq法RNA-seq是一种基于第二代高通量测序平台的转录组分析方法。相比DNA微阵列法,RNA-seq所需的RNA样品更少,无需相应物种的基因组序列作为参照,具有本底信号更低、准确性更高、重复性好等优点[48]。差异RNA-seq (differential RNA sequencing, dRNA-seq)方法可以精确鉴定基因的转录起始位点及5′非翻译区(5′ untranslated region, 5′UTR)序列[49-50]。Soppa团队利用dRNA-seq方法检测了指数期Hfx. volcanii的初始转录组,鉴定了转录本的转录起始位点并绘制出转录本的5′UTR图谱[51]。利用dRNA-seq所得到的转录组学数据的长度信息不足,该团队在后续工作中使用RNA-seq方法进一步确定了转录本的长度[52]。
3 嗜盐古菌的组成型启动子组成型启动子是指在该类启动子控制下,基因的表达水平基本处于恒定,不受反式作用因子的调节。在嗜盐古菌中,已经报道了包括PamyH、PphaR、Pfdx、Plsm以及Prpl137e在内的5种组成型启动子(表 2)。
| Promoter | Species | Features | Reference |
| PamyH | Har. hispanica | Active in both Har. hispanica and E. coli | [53-54] |
| PphaR | Hfx. mediterranei | PphaR is repressed by PhaR, and a strong and constitutive promoter is obtained by deleting its cis-regulatory region | [43] |
| Pfdx | Hfx. mediterranei | A constitutive and strong promoter | [55] |
| Plsm and Prpl137e | Hfx. mediterranei and Hfx. volcanii |
lsm overlaps with rpl137e, and Prpl137e is located in the ORF of lsm. Plsm initiates the co-transcription of lsm and rpl137e, while Prpl137e only initiates the transcription of rpl137e | [56-59] |
3.1 amyH启动子PamyH
来自西班牙盐盒菌(Haloarcula hispanica, Har. hispanica)的PamyH启动子负责amyH (编码α-淀粉酶)的转录,该启动子在Hfx. volcanii和E. coli中均具有活性[53-54]。PamyH启动子序列同时具有细菌启动子和古菌启动子的核心元件,揭示了该启动子在嗜盐古菌和大肠杆菌中同时具有启动子活性的机制[54]。陈向东团队基于PamyH构建了Hfx. volcanii和E. coli的穿梭表达载体pAJ,这为研究Hfx. volcanii和E. coli的保守蛋白的功能提供了有效的工具[26]。
3.2 phaR启动子PphaR许多细菌和古菌在营养不均衡的条件下,具有合成一种生物可降解塑料PHA的能力,PHA在胞内以颗粒形式存在,这一过程受到严格的调控[60-61]。PhaR是Hfx. mediterranei中参与PHA积累与颗粒形成的调节蛋白。phaR基因和颗粒结合蛋白编码基因phaP由同一个启动子PphaR进行转录,PhaR通过与PphaR结合抑制phaR和phaP的转录[43]。本文作者所在团队通过删除PphaR启动子的调控序列,将其改造为一个强组成型启动子,并应用于嗜盐古菌蛋白表达及基因回补等操作中。通过将强组成型启动子PphaR整合到Hfx. mediterranei的染色体上,增强番茄红素合成途径并提高了番茄红素产量[62]。同时,他们利用PphaR启动子在Hfx. volcanii中异源表达来自Hfx. mediterranei的蛋白,为后续实现该菌蛋白体外生化实验提供了可能[63-66]。此外,他们还利用该启动子表达了mini-CRISPR结构,分别在Har. hispanica和Hfx. mediterranei中建立了高效的基因编辑工具以及CRISPRi工具[67-68]。
3.3 fdx启动子Pfdx来自Hbt. salinarum和死海盐盒菌(Haloarcula marismortui, Har. marismortui)的铁氧还蛋白Fdx以[2Fe-2S]为氧化还原中心,其编码基因在各个时期的转录水平保持恒定,故其启动子Pfdx可用于衡量其它启动子的启动强度[55]。Gregor和Pfeifer在研究Hbt. salinarum的启动子PcA、PpA与Hfx. mediterranei的启动子PmcA的启动强度时,利用Pfdx和来自Hfx. alicantei的PbgaH作为对照启动子,Pfdx在指数期活性达到最高,PbgaH和3个gvpA启动子在稳定期活性最高[69]。Born与Pfeifer使用Pfdx启动子验证了改造后的嗜盐古菌报告基因gfp的稳定性表达[70]。本文作者所在团队利用启动子Pfdx实现了Har. marismortui中PhaC蛋白的过表达,确定了PhaC是PHA合酶的一个亚基[71]。
3.4 Plsm和Prpl37eSm和类Sm (Lsm)蛋白参与细胞中RNA的多种代谢途径[72-74],存在于真核生物和多数古菌中[73-76]。Lsm蛋白编码基因通常与核糖体蛋白编码基因rpl37e相邻或重叠[56]。在Hfx. mediterranei和Hfx. volcanii中,lsm均与其下游的rpl37e重叠,rpl37e的启动子Prpl37e在lsm的开放阅读框中。启动子Plsm通常负责起始lsm和rpl37e的共转录,而Prpl37e只负责起始rpl37e的转录,这保证了rpl37e较高的表达水平[56-59]。
4 诱导型启动子诱导型启动子的启动活性通常受到诱导物的激活或抑制,在其核心启动子序列上游一般包含被称为顺式作用元件的DNA序列,通过细胞外或细胞内信号从而激活或抑制下游基因的转录。直至目前,嗜盐古菌已报道了6种诱导型启动子,受糖/氧气/盐浓度诱导的气体囊泡蛋白(gas vesicle protein, gvp)基因启动子、热诱导启动子Phsp5、受果糖或戊糖诱导的系列启动子、受氧/光诱导的Pbop启动子、受磷或者K+诱导的启动子、受色氨酸诱导的Ptna启动子等(表 3)。
| Promoter | Species | Inducer | Features | Reference |
| Promoters of gvp | Hfx. mediterranei Hbt. salinarium |
Carbohydrates/ Oxygen/Salinity |
PD initiates the expression of GvpD and GvpE, while GvpE enhances the activity of PD and PA | [77-89] |
| Ptfb2 and Phsp5 | Hbt. salinarium | Heat | The expression of downstream genes is highly induced at high temperature (50 ℃) | [90-91] |
| PglpR-pfkB, PkdgK1, PPTS, etc. | Hfx. volcanii Hfx. mediterranei |
Fructose | PglpR-pfkB and PkdgK1 are repressed by GlpR when glycerol is used as the sole carbon source; this repression is relieved in the presence of fructose or glucose; GlpR activates PPTS | [92-93] |
| PxacB, PxacD, PxacEA, PxacF, and PxacR | Hfx. volcanii | Xylose and arabinose | XacR functions as the activator of PxacB, PxacD, PxacEA, and PxacF, and the repressor of PxacR, respectively | [94-95] |
| Pbop | Hbt. halobium | Hypoxia/Light | Pbop is activated by Bat upon exposure to hypoxia or light | [96-98] |
| Ppst | Hbt. salinarium | Phosphorus | Ppst2 is about 10 times stronger than Ppst1 under excess phosphorus conditions, while Ppst1 is about 10 times stronger than Ppst2 under phosphorus limitation | [99] |
| Pkdp | Hbt. salinarium | K+ | Pkdp is repressed by high concentration of K+ | [100-102] |
| Ptna | Hfx. volcanii | Tryptophan | Tryptophan is an efficient and stringent inducer of Ptna | [33] |
| PnasABC | Hfx. mediterranei | Nitrogen starvation | Lrp/AsnC are potential transcriptional factors involved in regulating nasABC transcription | [103-104] |
4.1 gvp基因启动子
某些嗜盐古菌(如Hbt. salinarium和Hfx. mediterranei)拥有编码气泡的基因,这些基因所编码的蛋白质,可以形成充满气体的蛋白质纳米囊泡[105]。气体囊泡增加了细胞的浮力,使其能够在水体中垂直迁移到生长条件最佳的区域。分析发现共有14个gvp基因参与嗜盐古菌气体囊泡的形成,它们以2个基因簇的形式(gvpACNO及gvpDEFGHIJKLM)相反地排列在基因组中[77]。其中,Hfx. mediterranei存在单一拷贝的14个gvp基因,被命名为mc-vac,而Hbt. salinarium存在2个拷贝的14个gvp基因,分别位于染色体及大质粒上,被命名为c-vac与p-vac[78]。气体囊泡的形成受到糖、氧气和盐浓度的影响[79-82]。在好氧条件下,随着培养基中葡萄糖、麦芽糖或者蔗糖浓度的增加,Hfx. mediterranei中气体囊泡的数量持续减少;而在厌氧条件下,菌体内缺乏气体囊泡,gvp基因的转录水平降低了10倍。
Hbt. salinarium的p-vac的14个gvp基因由4个启动子PA、PD、PF和PO负责转录,而Hbt. salinarium的c-vac和Hfx. mediterranei的mc-vac的14个gvp基因由PA和PD负责转录(图 2)[83-84]。在p-vac中,活性最强的PA负责GvpA和GvpC的表达[85];PD负责GvpD和GvpE的表达,GvpE是PD和PA的转录激活因子[24, 86],GvpD通过与GvpE形成复合物抑制GvpE蛋白对PD和PA转录的激活,最终抑制气体囊泡的形成[87-88]。PF和PO不受GvpD和GvpE的调控[78]。PA与PD启动子上游各存在一段上游激活序列(upstream activating sequence, UAS, UASA和UASD),GvpE通过与UASA和UASD结合而激活PD和PA的转录活性。p-vac和mc-vac的UASA与UASD存在部分重叠序列,且二者紧邻PA与PD的BRE元件,因此GvpE蛋白通过结合UASA与UASD同时增强PA与PD的活性[89]。而c-vac的UASD和PD间多间隔了22 nt,这可能导致了GvpE可以激活PA活性,而无法增强c-vac中PD的活性[88] (图 2)。
4.2 高温诱导启动子Ptfb2和Phsp5
热休克应答是在三域生命中普遍存在的生理现象,科学家从响应热刺激的热休克蛋白编码基因的启动子中已经发现众多受温度调控的启动子[106-108]。其中,嗜盐古菌中报道的热休克蛋白基因启动子,包括高温诱导的tfb2、cct1、hsp5启动子和低温诱导的cspD2启动子[90-91, 109-110]。
Hfx. volcanii中已有多个编码转录因子的tfb基因被鉴定,其中tfb2在转录水平上特异性响应热激[90]。60 ℃诱导15 min,tfb2的mRNA水平提高了8倍,这说明tfb2的核心启动子具有热响应特性。本文作者所在团队在解析Hbt. salinarum热休克基因hsp5的热诱导转录调控机制时发现,该基因主要通过Phsp5的BRE和TATA盒进行调控,而删除BRE上游序列和突变TATA盒下游序列对该启动子基础转录和热诱导活性均影响甚微[91]。DNA结合实验表明,来自Halobacterium的TFBb和来自Haloferax的TFB2均可与Phsp5特异性结合激活了hsp5的转录,且在热诱导对应的高温条件下其结合活性更强,从而实现了hsp5的热诱导表达。该项工作为古菌基础转录因子直接主导环境响应基因表达提供了一个范例[91]。
4.3 糖代谢相关基因启动子某些嗜盐古菌可以利用多种碳源,包括甘油、果糖、葡萄糖、木糖和几丁质等[92, 111]。Hfx. volcanii的多种六碳糖代谢都受到GlpR蛋白的调控,该蛋白属于具有螺旋-转角-螺旋(helix-turn-helix, HTH)结构域的DeoR调控蛋白家族。戊糖分解相关基因xac的启动子则受到转录因子XacR的调控。
GlpR蛋白可抑制Hfx. volcanii中葡萄糖和果糖代谢相关基因的转录[93]。Hfx. volcanii的葡萄糖和果糖代谢的关键编码基因kdgK1和pfkB的转录,分别受葡萄糖或果糖诱导。GlpR是pfkB和kdgK1的转录抑制子,glpR和pfkB的转录由同一个启动子PglpR-pfkB启动,GlpR通过与PglpR-pfkB结合抑制glpR和pfkB的转录。细胞摄取的果糖被转化为F-1-P,F-1-P是GlpR的特异性配体,通过结合GlpR使其与PglpR-pfkB的结合能力减弱,解除其对glpR-pfkB、fba等果糖代谢基因转录的抑制,最终提高相关基因的转录水平[92],促进果糖的吸收利用。Hfx. mediterranei的GlpR是糖磷酸转移酶系统基因簇PTS表达的激活因子[47],通过直接与PPTS启动子序列的特异性结合实现对PTS基因簇的调控。此时,果糖诱导野生菌的PTS基因簇和glpR-fruK操纵子转录水平的上调表达,而在突变株ΔglpR中,这一诱导现象消失。
Hfx. volcanii的戊糖分解调控基因簇包括xacF、xacB、xacD和xacEA,它们的表达受底物d-木糖或者l-阿拉伯糖的诱导,XacR是上述基因的转录激活因子,由xacR基因编码,属于IclR转录调控因子家族[94-95]。XacR同时也是其自身编码基因xacR的阻遏蛋白。xacR基因转录起始位点前富含AT的回文序列是XacR的阻遏位点,戊糖可以引发配体与XacR效应位点结合以解除XacR对xacR的抑制,从而开启相关基因的转录以进行木糖代谢。
4.4 细菌视蛋白基因启动子Pbop原核生物的视黄醛蛋白细菌视紫红质(bacteriorhodopsin, BR)由bop基因编码,是嗜盐古菌紫膜中的光驱动质子泵[112]。bop所在基因簇还包括bat、brp和bip基因,其中bat基因编码一种反式作用因子,该因子又被称为双效应因子,在低氧胁迫或高光强度时均激活bop的转录。Bat蛋白的环磷酸鸟苷调节的磷酸二酯酶(cGMP-regulated phosphodiesterases, PDEs),鱼腥藻腺苷酸环化酶(Anabaena adenylyl cyclases, ACs),E. coli转录激活子FhlA (transcription activator FhlA, GAF)和周期时钟蛋白-芳香烃受体-Sim蛋白(period clock protein-aryl hydrocarbon receptor-single minded protein, Per-ARNT-Sim, PAS)/PAS相关的C末端(PAS-associated C-terminal, PAC)结构域分别用于响应光与低氧胁迫,其HTH基序负责与bop启动子上游调控序列结合[96-97],通过改变DNA的超螺旋结构以实现对bop转录的调控[98]。
4.5 转运蛋白相关基因启动子ABC转运复合物磷酸转运系统(phosphate transport system, PST)是一种对磷具有高亲和力的主动转运系统,负责将磷特异性地转入细胞[113]。Hbt. salinarium的基因组拥有编码该系统的2个操纵子(pst1和pst2),均为多顺反子。在磷饱和的条件下,pst1操纵子具有2个转录起始位点(transcription start site, TSS) TSS-1与TSS-2,利用率之比约为4:1,在无磷的培养条件下,仅能检测到TSS-1作为转录起始位点。pst2转录本只具有一个转录起始位点。以TSS-1为起始位点的pst1转录本比以TSS-2为起始位点的转录本翻译效率更高。因此,在限磷条件下,pst1短转录本在磷元素转运过程中发挥重要的生理功能[99]。pst操纵子的转录由Ppst驱动,Ppst磷依赖性的调控元件均含有AT富集序列(ATATATA),被称为P盒,它是保持Ppst活性的必要元件[99]。
Hbt. salinarium的基因组编码ATP依赖性的高亲和力K+摄取系统Kdp,该系统包含KdpFABC和ATPase,分别由结构基因kdpFABC与kdpQ编码。基因簇kdpFABCQ以单个多顺反子mRNA形式进行转录[100-101]。Pkdp的启动强度随着培养基中K+浓度的升高而降低,而诱导后Pkdp的启动强度可增加50倍。Kixmüller与Greie利用Pkdp对磷的敏感性及诱导性,构建了受K+浓度诱导的蛋白表达系统[102]。
4.6 受色氨酸诱导的Ptna在许多生物中,色氨酸的合成基因trp通常受到严格调控。在Hfx. volcanii中,色氨酸合成的相关酶由trpCBA与trpDFEG2个基因簇编码,它们的表达分别由其各自共转录启动子驱动,在色氨酸浓度升高时它们的转录被抑制[114-115]。tna是Hfx. volcanii中的色氨酸酶(tryptophanase)编码基因,受色氨酸诱导表达[33]。Ptna是一个本底表达水平低、且能被色氨酸高效诱导的启动子,在Hfx. volcanii中可实现基因的可诱导差异表达。整合到染色体上的Ptna也可被色氨酸严谨而高效地诱导。Lund团队还在Hfx. volcanii中利用Ptna构建了cct1缺失的条件致死基因突变株,研究了分子伴侣蛋白CCT1的功能[33]。
4.7 硝酸盐诱导的PnasABCHfx. mediterranei的硝酸还原酶(nitrate reductase enzyme, Nas)由nasA编码,而nasA与nasB、nasC由启动子PnasABC启动共转录,转录过程受到硝酸盐诱导[103]。Pastor-Soler等使用β-半乳糖苷酶作为报告基因,通过对PnasABC启动子区域的定点诱变确认转录因子结合区域,然后通过pull-down实验结合质谱法在体外鉴定了PnasABC的转录调控因子,该蛋白属于Lrp/AsnC家族,但其体内的分子作用机制尚不明确[104]。
5 嗜盐古菌启动子的应用 5.1 目标蛋白过表达在嗜盐古菌的很多研究中,会通过将目标蛋白带上纯化标签,对其进行过表达,然后利用纯化技术得到纯度较高的蛋白,从而为体外生化实验提供实验材料,在这个过程中,就需要利用强启动子尽可能提高目标蛋白的表达量。本文作者所在团队在Hfx. mediterranei的多种未知蛋白功能(如PHA解聚酶PhaZh1[66]、PPS和PYK[63])的研究工作中,使用强启动子PphaR过表达带有His标签的目的蛋白,然后对纯化后的蛋白进行了体外酶活功能的研究。
5.2 基因回补在对基因功能的体内遗传学研究中,通常需要基因回补操作,以回补敲除基因后的表型,此时可利用诱导型启动子通过调整诱导物浓度而方便调整回补后的基因表达水平。Aharonowitz团队在对Hfx. volcanii中γ-谷氨酰半胱氨酸连接酶编码基因gshA的鉴定工作中,发现敲除gshA的菌株中缺少γ-谷氨酰半胱氨酸(γ-glutamylcysteine, γGC)[116]。为验证γGC的缺乏是由gshA的缺失引起的,研究者在ΔgshA的菌株中回补了由Ptna控制的gshA,最终在回补菌株提取物中检测到了γGC,由此确定了Hfx. volcanii中gshA的功能为合成并积累γGC。
5.3 代谢通路重构为了提高嗜盐古菌在工业生产中的产物产量或合成原本不能合成的高值产品,可通过对嗜盐古菌代谢通路的改造甚至重构来实现。启动子为代谢通路的构建提供了重要的基因表达调控手段。
本文作者所在团队对Hfx. mediterranei的番茄红素合成途径进行了代谢工程改造[62]。他们首先鉴定了番茄红素合成途径中限速酶是八氢番茄红素合酶(CrtB),该酶催化牻牛儿基牻牛儿基焦磷酸(GGPP)生成八氢番茄红素。在此基础上利用强启动子PphaR过表达自身CrtB,并利用该启动子异源表达了来自近缘属种的CrtB和八氢番茄红素去饱和酶CrtI,使番茄红素产量变得更高。最终,经代谢工程改造的Hfx. mediterranei菌株在摇瓶发酵中每克干细胞可以生产(119.25±0.55) mg番茄红素。
6 展望随着嗜盐古菌相关工作大量展开,可应用于嗜盐古菌的研究工具与技术也得到了不断地扩充,但相比于传统模式微生物,嗜盐古菌启动子的研究应用仍处于初级阶段,具有较大的发展空间。在近年来备受关注的合成生物学领域,启动子是实现基因精细表达调控的重要核心元件之一,而标准化启动子工具箱正是嗜盐古菌所缺乏的。
将来有关该方面工作有望在以下几方面取得进展:
(1) 更多天然启动子的发现与鉴定。利用现有技术,如RNA-Seq测序等,寻找并开发更多不同强度的组成型和诱导型启动子。随着测序技术的发展,期待有更便捷的方法对转录组进行扫描、对启动子进行鉴定。此外,嗜盐古菌的基因组GC含量高、转录本多数无5′UTR,期待根据这些特点开发出研究嗜盐古菌启动子的特异性方法与策略。
(2) 启动子活性的预测和校正。组学数据配合机器学习算法的使用可以帮助我们预测启动子的活性强度,但由于不同菌种间的差异会对启动子活性预测的通用性造成困难,因此还需要相应的实验对启动子活性进行校正。这项工作可以促进嗜盐古菌合成生物学中启动子元件的开发,使代谢工程改造的结果可以被更准确地预测。
(3) 启动子的分子机制解析和人工改造。嗜盐古菌的启动子可能具有全新的分子机制,尤其是针对诱导型启动子,其序列结构、调控机制、效应蛋白的解析是了解启动子并对其进行人工改造的前提。在清楚解析启动子分子机制后,就可以通过对启动子各结构元件的突变、替换和组装,实现对启动子进行理性改造,达到增强或减弱启动子的活性,降低启动子的表达渗漏,以及构建多种诱导类型的启动子,获得更多且更符合启动强度需求的启动子元件的目的。
此外,启动子在与核糖开关、群体感应信号等其他转录调控元件的组合后,也将得到更多正交的转录调控系统[117]。例如,将诱导型启动子与核糖开关串联的转录调控系统,可以同时使用2种效应物对转录进行更精准的控制。目前在嗜盐古菌Hfx. volcanii中,已报道了一种使用茶碱作为效应物的核糖开关被应用于控制gvpE的转录[118]。
相比细菌,嗜盐古菌启动子结构更接近于真核细胞。但目前已发现极少数的嗜盐古菌启动子同时具有古菌、细菌和真核细胞启动子的典型序列特征,这类启动子在古菌、细菌甚至真核细胞中均具有启动子功能,如本文3.1中介绍的PamyH启动子。该发现为构建双功能或多功能表达载体提供了全新的启动子元件,我们期待能够在嗜盐古菌中开发出更多可同时应用到古菌、细菌和真核细胞中的启动子。同时,我们还期待对嗜盐古菌启动子的改造工作可以对嗜盐古菌基因表达调控实现更有效的控制,也相信嗜盐古菌启动子元件的进一步开发将扩充嗜盐古菌合成生物学研究的工具箱,从而加速嗜盐古菌底盘细胞的改造、优化和升级,促进嗜盐古菌遗传代谢和合成生物学等多领域的研究。
| [1] | WOESE CR, KANDLER O, WHEELIS ML. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proceedings of the National Academy of Sciences of the United States of America, 1990, 87(12): 4576-4579. DOI:10.1073/pnas.87.12.4576 |
| [2] | BAKER BJ, de ANDA V, SEITZ KW, DOMBROWSKI N, SANTORO AE, LLOYD KG. Diversity, ecology and evolution of Archaea. Nature Microbiology, 2020, 5(7): 887-900. DOI:10.1038/s41564-020-0715-z |
| [3] | ISHINO Y, KRUPOVIC M, FORTERRE P. History of CRISPR-cas from encounter with a mysterious repeated sequence to genome editing technology. Journal of Bacteriology, 2018, 200(7): e00580-e00517. |
| [4] | PFEIFER K, ERGAL İ, KOLLER M, BASEN M, SCHUSTER B, RITTMANN SKM R. Archaea biotechnology. Biotechnology Advances, 2021, 47: 107668. DOI:10.1016/j.biotechadv.2020.107668 |
| [5] |
SHEN P. Research of haloarchaeal molecular genetics[C]. The 7th Seminar of the Genetics Society of HuBei Province, Wuchang China, F, 2004 (in Chinese). 沈萍. 极端嗜盐古菌的分子遗传学研究. 湖北省遗传学会第七次代表大会暨学术讨论会, 中国, 武昌, F, 2004. |
| [6] |
XIANG H. Research of haloarchaeal molecular genetics and physiological metabolism[C]. The 8th Seminar of the Genetics Society of China, Chongqing China, F, 2008 (in Chinese). 向华. 极端嗜盐古菌分子遗传与生理代谢研究。中国遗传学会第八次代表大会暨学术讨论会, 中国, 重庆, F, 2008. |
| [7] | LIFTON RP, GOLDBERG ML, KARP RW, HOGNESS DS. The organization of the histone genes in Drosophila melanogaster: functional and evolutionary implications. Cold Spring Harbor Symposia on Quantitative Biology, 1978, 42 Pt 2: 1047-1051. |
| [8] | LAGRANGE T, KAPANIDIS AN, TANG H, REINBERG D, EBRIGHT RH. New core promoter element in RNA polymerase II-dependent transcription: sequence-specific DNA binding by transcription factor IIB. Genes & Development, 1998, 12(1): 34-44. |
| [9] | SMALE ST, BALTIMORE D. The "initiator" as a transcription control element. Cell, 1989, 57(1): 103-113. DOI:10.1016/0092-8674(89)90176-1 |
| [10] | REITER WD, HÜDEPOHL U, ZILLIG W. Mutational analysis of an archaebacterial promoter: essential role of a TATA box for transcription efficiency and start-site selection in vitro. Proceedings of the National Academy of Sciences of the United States of America, 1990, 87(24): 9509-9513. DOI:10.1073/pnas.87.24.9509 |
| [11] | HAUSNER W, FREY G, THOMM M. Control regions of an archaeal gene. A TATA box and an initiator element promote cell-free transcription of the tRNA (Val) gene of Methanococcus vannielii. Journal of Molecular Biology, 1991, 222(3): 495-508. DOI:10.1016/0022-2836(91)90492-O |
| [12] | BELL SD, JACKSON SP. Transcription and translation in Archaea: a mosaic of eukaryal and bacterial features. Trends in Microbiology, 1998, 6(6): 222-228. DOI:10.1016/S0966-842X(98)01281-5 |
| [13] | BELL SD, JACKSON SP. Mechanism and regulation of transcription in Archaea. Current Opinion in Microbiology, 2001, 4(2): 208-213. DOI:10.1016/S1369-5274(00)00190-9 |
| [14] | QURESHI SA, JACKSON SP. Sequence-specific DNA binding by the S. shibatae TFIIB homolog, TFB, and its effect on promoter strength. Molecular Cell, 1998, 1(3): 389-400. DOI:10.1016/S1097-2765(00)80039-8 |
| [15] | BELL SD, KOSA PL, SIGLER PB, JACKSON SP. Orientation of the transcription preinitiation complex in Archaea. Proceedings of the National Academy of Sciences of the United States of America, 1999, 96(24): 13662-13667. DOI:10.1073/pnas.96.24.13662 |
| [16] | LANGER D, HAIN J, THURIAUX P, ZILLIG W. Transcription in Archaea: similarity to that in eucarya. Proceedings of the National Academy of Sciences of the United States of America, 1995, 92(13): 5768-5772. DOI:10.1073/pnas.92.13.5768 |
| [17] | DEDECKER BS, O'BRIEN R, FLEMING PJ, GEIGER JH, JACKSON SP, SIGLER PB. The crystal structure of a hyperthermophilic archaeal TATA-box binding protein. Journal of Molecular Biology, 1996, 264(5): 1072-1084. DOI:10.1006/jmbi.1996.0697 |
| [18] | LITTLEFIELD O, KORKHIN Y, SIGLER PB. The structural basis for the oriented assembly of a TBP/TFB/promoter complex. Proceedings of the National Academy of Sciences of the United States of America, 1999, 96(24): 13668-13673. DOI:10.1073/pnas.96.24.13668 |
| [19] | MEINHART A, BLOBEL J, CRAMER P. An extended winged helix domain in general transcription factor E/IIEα. Journal of Biological Chemistry, 2003, 278(48): 48267-48274. DOI:10.1074/jbc.M307874200 |
| [20] | NAJI S, GRÜNBERG S, THOMM M. The RPB7 orthologue E′ is required for transcriptional activity of a reconstituted archaeal core enzyme at low temperatures and stimulates open complex formation. The Journal of Biological Chemistry, 2007, 282(15): 11047-11057. DOI:10.1074/jbc.M611674200 |
| [21] | HAUSNER W, THOMM M. Events during initiation of archaeal transcription: open complex formation and DNA-protein interactions. Journal of Bacteriology, 2001, 183(10): 3025-3031. DOI:10.1128/JB.183.10.3025-3031.2001 |
| [22] | GEHRING AM, WALKER JE, SANTANGELO TJ. Transcription regulation in Archaea. Journal of Bacteriology, 2016, 198(14): 1906-1917. DOI:10.1128/JB.00255-16 |
| [23] | BYLINO OV, IBRAGIMOV AN, SHIDLOVSKII YV. Evolution of regulated transcription. Cells, 2020, 9(7): 1675. DOI:10.3390/cells9071675 |
| [24] | HOLMES ML, SCOPES RK, MORITZ RL, SIMPSON RJ, Englert C, Pfeifer F, Dyall-Smith ML. Purification and analysis of an extremely halophilic beta-galactosidase from Haloferax alicantei. Biochimica et Biophysica Acta, 1997, 1337(2): 276-286. DOI:10.1016/S0167-4838(96)00174-4 |
| [25] | GREGOR D, PFEIFER F. Use of a halobacterial bgaH reporter gene to analyse the regulation of gene expression in halophilic Archaea. Microbiology: Reading, England, 2001, 147(Pt 7): 1745-1754. |
| [26] | LV J, WANG S, ZENG C, HUANG YP, CHEN XD. Construction of a shuttle expression vector with a promoter functioning in both halophilic Archaea and Bacteria. FEMS Microbiology Letters, 2013, 349(1): 9-15. |
| [27] |
HUANG YP, DUAN ZH, XIONG Y, GUO PY, SHEN P. Sequence characteristic and function of DNA fragments active as bacteria promoter from Archaea. Wuhan University Journal: Natural Science Edition, 2001, 47(4): 456-462.
(in Chinese) 黄玉屏, 段珍红, 熊音, 郭培懿, 沈萍. 盐生盐杆菌启动子DNA片段的特征序列及其功能分析. 武汉大学学报: 理学版, 2001, 47(4): 456-462. |
| [28] | STOLT P, ZILLIG W. Antisense RNA mediates transcriptional processing in an archaebacterium, indicating a novel kind of RNase activity. Molecular Microbiology, 1993, 7(6): 875-882. DOI:10.1111/j.1365-2958.1993.tb01178.x |
| [29] | TANG SL, NUTTALL S, NGUI K, FISHER C, LOPEZ P, DYALL-SMITH M. HF2: a double-stranded DNA tailed haloarchaeal virus with a mosaic genome. Molecular Microbiology, 2002, 44(1): 283-296. DOI:10.1046/j.1365-2958.2002.02890.x |
| [30] | DYALL-SMITH M, TANG SL, RUSS B, CHIANG PW, PFEIFFER F. Comparative genomics of two new HF1-like haloviruses. Genes, 2020, 11(4): 405. DOI:10.3390/genes11040405 |
| [31] | RUSS B, PFEIFFER F, DYALL-SMITH M. Halovirus HF2 intergenic repeat sequences carry promoters. Viruses, 2021, 13(12): 2388. DOI:10.3390/v13122388 |
| [32] | BITAN-BANIN G, ORTENBERG R, MEVARECH M. Development of a gene knockout system for the halophilic archaeon Haloferax volcanii by use of the pyrE gene. Journal of Bacteriology, 2003, 185(3): 772-778. DOI:10.1128/JB.185.3.772-778.2003 |
| [33] | LARGE A, STAMME C, LANGE C, DUAN ZH, ALLERS T, SOPPA J, LUND PA. Characterization of a tightly controlled promoter of the halophilic archaeon Haloferax volcanii and its use in the analysis of the essential cct1 gene. Molecular Microbiology, 2007, 66(5): 1092-1106. DOI:10.1111/j.1365-2958.2007.05980.x |
| [34] | SCHWEITZER BI, DICKER AP, BERTINO JR. Dihydrofolate reductase as a therapeutic target. FASEB Journal: Official Publication of the Federation of American Societies for Experimental Biology, 1990, 4(8): 2441-2452. DOI:10.1096/fasebj.4.8.2185970 |
| [35] | ROSENSHINE I, ZUSMAN T, WERCZBERGER R, MEVARECH M. Amplification of specific DNA sequences correlates with resistance of the archaebacterium Halobacterium volcanii to the dihydrofolate reductase inhibitors trimethoprim and methotrexate. Molecular and General Genetics MGG, 1987, 208(3): 518-522. DOI:10.1007/BF00328149 |
| [36] | ZUSMAN T, ROSENSHINE I, BOEHM G, JAENICKE R, LESKIW B, MEVARECH M. Dihydrofolate reductase of the extremely halophilic archaebacterium Halobacterium volcanii. Journal of Biological Chemistry, 1989, 264(32): 18878-18883. DOI:10.1016/S0021-9258(19)47238-4 |
| [37] | DANNER S, SOPPA J. Characterization of the distal promoter element of halobacteria in vivo using saturation mutagenesis and selection. Molecular Microbiology, 1996, 19(6): 1265-1276. DOI:10.1111/j.1365-2958.1996.tb02471.x |
| [38] | TSIEN RY. The green fluorescent protein. Annual Review of Biochemistry, 1998, 67: 509-544. DOI:10.1146/annurev.biochem.67.1.509 |
| [39] | CHALFIE M, TU Y, EUSKIRCHEN G, WARD WW, PRASHER DC. Green fluorescent protein as a marker for gene expression. Science, 1994, 263(5148): 802-805. DOI:10.1126/science.8303295 |
| [40] | CRAMERI A, WHITEHORN EA, TATE E, STEMMER WPC. Improved green fluorescent protein by molecular evolution using DNA shuffling. Nature Biotechnology, 1996, 14(3): 315-319. DOI:10.1038/nbt0396-315 |
| [41] | DAVIS SJ, VIERSTRA RD. Soluble, highly fluorescent variants of green fluorescent protein (GFP) for use in higher plants. Plant Molecular Biology, 1998, 36(4): 521-528. DOI:10.1023/A:1005991617182 |
| [42] | REUTER CJ, MAUPIN-FURLOW JA. Analysis of proteasome-dependent proteolysis in Haloferax volcanii cells, using short-lived green fluorescent proteins. Applied and Environmental Microbiology, 2004, 70(12): 7530-7538. DOI:10.1128/AEM.70.12.7530-7538.2004 |
| [43] | CAI SF, CAI L, ZHAO DH, LIU GM, HAN J, ZHOU J, XIANG H. A novel DNA-binding protein, PhaR, plays a central role in the regulation of polyhydroxyalkanoate accumulation and granule formation in the haloarchaeon Haloferax mediterranei. Applied and Environmental Microbiology, 2015, 81(1): 373-385. DOI:10.1128/AEM.02878-14 |
| [44] | FODOR SPA, RAVA RP, HUANG XC, PEASE AC, HOLMES CP, ADAMS CL. Multiplexed biochemical assays with biological chips. Nature, 1993, 364(6437): 555-556. DOI:10.1038/364555a0 |
| [45] | SCHENA M, SHALON D, DAVIS RW, BROWN PO. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science, 1995, 270(5235): 467-470. DOI:10.1126/science.270.5235.467 |
| [46] | STRILLINGER E, GRÖTZINGER SW, ALLERS T, EPPINGER J, WEUSTER-BOTZ D. Production of halophilic proteins using Haloferax volcanii H1895 in a stirred-tank bioreactor. Applied Microbiology and Biotechnology, 2016, 100(3): 1183-1195. DOI:10.1007/s00253-015-7007-1 |
| [47] | CAI L, CAI SF, ZHAO DH, WU JH, WANG L, LIU XQ, LI M, HOU J, ZHOU J, LIU JF, HAN J, XIANG H. Analysis of the transcriptional regulator GlpR, promoter elements, and posttranscriptional processing involved in fructose-induced activation of the phosphoenolpyruvate-dependent sugar phosphotransferase system in Haloferax mediterranei. Applied and Environmental Microbiology, 2014, 80(4): 1430-1440. DOI:10.1128/AEM.03372-13 |
| [48] | WANG Z, GERSTEIN M, SNYDER M. RNA-Seq: a revolutionary tool for transcriptomics. Nature Reviews Genetics, 2009, 10(1): 57-63. DOI:10.1038/nrg2484 |
| [49] | SHARMA CM, HOFFMANN S, DARFEUILLE F, REIGNIER J, FINDEIß S, SITTKA A, CHABAS S, REICHE K, HACKERMÜLLER J, REINHARDT R, STADLER PF, VOGEL J. The primary transcriptome of the major human pathogen Helicobacter pylori. Nature, 2010, 464(7286): 250-255. DOI:10.1038/nature08756 |
| [50] | SHARMA CM, Vogel J. Differential RNA-seq: The approach behind and the biological insight gained. Current Opinion in Microbiology, 2014, 19: 97-105. DOI:10.1016/j.mib.2014.06.010 |
| [51] | BABSKI J, HAAS KA, NÄTHER-SCHINDLER D, PFEIFFER F, FÖRSTNER KU, HAMMELMANN M, HILKER R, BECKER A, SHARMA CM, MARCHFELDER A, SOPPA J. Genome-wide identification of transcriptional start sites in the haloarchaeon Haloferax volcanii based on differential RNA-Seq (dRNA-Seq). BMC Genomics, 2016, 17(1): 629. DOI:10.1186/s12864-016-2920-y |
| [52] | LAASS S, MONZON VA, KLIEMT J, HAMMELMANN M, PFEIFFER F, FÖRSTNER KU, SOPPA J. Characterization of the transcriptome of Haloferax volcanii, grown under four different conditions, with mixed RNA-Seq. PLoS One, 2019, 14(4): e0215986. DOI:10.1371/journal.pone.0215986 |
| [53] | HUTCHEON GW, VASISHT N, Bolhuis A. Characterisation of a highly stable α-amylase from the halophilic archaeon Haloarcula hispanica. Extremophiles, 2005, 9(6): 487-495. DOI:10.1007/s00792-005-0471-2 |
| [54] | ZENG C, ZHAO YZ, CUI CZ, ZHANG H, ZHU JY, TANG XF, SHEN P, HUANG YP, CHEN XD. Characterization of the Haloarcula hispanica amyH gene promoter, an archaeal promoter that confers promoter activity in Escherichia coli. Gene, 2009, 442(1/2): 1-7. |
| [55] | PFEIFER F, GRIFFIG J, OESTERHELT D. The fdx gene encoding the [2Fe-2S] ferredoxin of Halobacterium salinarium (H. halobium). Molecular and General Genetics MGG, 1993, 239(1): 66-71. |
| [56] | MURA C, RANDOLPH PS, PATTERSON J, COZEN AE. Archaeal and eukaryotic homologs of Hfq: a structural and evolutionary perspective on Sm function. RNA Biology, 2013, 10(4): 636-651. DOI:10.4161/rna.24538 |
| [57] | PAYÁ G, BAUTISTA V, CAMACHO M, BONETE MJ, ESCLAPEZ J. Functional analysis of Lsm protein under multiple stress conditions in the extreme haloarchaeon Haloferax mediterranei. Biochimie, 2021, 187: 33-47. DOI:10.1016/j.biochi.2021.05.002 |
| [58] | FISCHER S, BENZ J, SPÄTH B, MAIER LK, STRAUB J, GRANZOW M, RAABE M, URLAUB H, HOFFMANN J, BRUTSCHY B, ALLERS T, SOPPA J, MARCHFELDER A. The archaeal Lsm protein binds to small RNAs. The Journal of Biological Chemistry, 2010, 285(45): 34429-34438. DOI:10.1074/jbc.M110.118950 |
| [59] | MAIER LK, BENZ J, FISCHER S, ALSTETTER M, JASCHINSKI K, HILKER R, BECKER A, ALLERS T, SOPPA J, MARCHFELDER A. Deletion of the Sm1 encoding motif in the lsm gene results in distinct changes in the transcriptome and enhanced swarming activity of Haloferax cells. Biochimie, 2015, 117: 129-137. DOI:10.1016/j.biochi.2015.02.023 |
| [60] | JENDROSSEK D, PFEIFFER D. New insights in the formation of polyhydroxyalkanoate granules (carbonosomes) and novel functions of poly(3-hydroxybutyrate). Environmental Microbiology, 2014, 16(8): 2357-2373. DOI:10.1111/1462-2920.12356 |
| [61] | PÖTTER M, Steinbüchel A. Poly(3-hydroxybutyrate) granule-associated proteins: impacts on poly(3-hydroxybutyrate) synthesis and degradation. Biomacromolecules, 2005, 6(2): 552-560. DOI:10.1021/bm049401n |
| [62] | ZUO ZQ, XUE Q, ZHOU J, ZHAO DH, HAN J, XIANG H. Engineering Haloferax mediterranei as an efficient platform for high level production of lycopene. Frontiers in Microbiology, 2018, 9: 2893. DOI:10.3389/fmicb.2018.02893 |
| [63] | CHEN JY, MITRA R, ZHANG SJ, ZUO ZQ, LIN L, ZHAO DH, XIANG H, HAN J. Unusual phosphoenolpyruvate (PEP) synthetase-like protein crucial to enhancement of polyhydroxyalkanoate accumulation in Haloferax mediterranei revealed by dissection of PEP-pyruvate interconversion mechanism. Applied and Environmental Microbiology, 2019, 85(19): e00984-e00919. |
| [64] | CHEN JY, MITRA R, XIANG H, HAN J. Deletion of the pps-like gene activates the cryptic phaC genes in Haloferax mediterranei. Applied Microbiology and Biotechnology, 2020, 104(22): 9759-9771. DOI:10.1007/s00253-020-10898-0 |
| [65] | LIU GM, CAI SF, HOU J, ZHAO DH, HAN J, ZHOU J, XIANG H. Enoyl-CoA hydratase mediates polyhydroxyalkanoate mobilization in Haloferax mediterranei. Scientific Reports, 2016, 6: 24015. DOI:10.1038/srep24015 |
| [66] | LIU GM, HOU J, CAI SF, ZHAO DH, CAI L, HAN J, ZHOU J, XIANG H. A patatin-like protein associated with the polyhydroxyalkanoate (PHA) granules of Haloferax mediterranei acts as an efficient depolymerase in the degradation of native PHA. Applied and Environmental Microbiology, 2015, 81(9): 3029-3038. DOI:10.1128/AEM.04269-14 |
| [67] | CHENG FY, GONG LY, ZHAO DH, YANG HB, ZHOU J, LI M, XIANG H. Harnessing the native type I-B CRISPR-Cas for genome editing in a polyploid archaeon. Journal of Genetics and Genomics, 2017, 44(11): 541-548. DOI:10.1016/j.jgg.2017.09.010 |
| [68] | LIN L, CHEN JY, MITRA R, GAO QX, CHENG FY, XU T, ZUO ZQ, XIANG H, HAN J. Optimising PHBV biopolymer production in haloarchaea via CRISPRi-mediated redirection of carbon flux. Communications Biology, 2021, 4: 1007. DOI:10.1038/s42003-021-02541-z |
| [69] | GREGOR D, PFEIFER F. In vivo analyses of constitutive and regulated promoters in halophilic Archaea. Microbiology: Reading, England, 2005, 151(Pt 1): 25-33. |
| [70] | BORN J, PFEIFER F. Improved GFP variants to study gene expression in haloarchaea. Frontiers in Microbiology, 2019, 10: 1200. DOI:10.3389/fmicb.2019.01200 |
| [71] | HAN J, LU QH, ZHOU LG, ZHOU J, XIANG H. Molecular characterization of the phaECHm genes, required for biosynthesis of poly(3-hydroxybutyrate) in the extremely halophilic archaeon Haloarcula marismortui. Applied and Environmental Microbiology, 2007, 73(19): 6058-6065. DOI:10.1128/AEM.00953-07 |
| [72] | SÉRAPHIN B. Sm and Sm-like proteins belong to a large family: identification of proteins of the U6 as well as the U1, U2, U4 and U5 snRNPs. The EMBO Journal, 1995, 14(9): 2089-2098. DOI:10.1002/j.1460-2075.1995.tb07200.x |
| [73] | SCOFIELD DG, LYNCH M. Evolutionary diversification of the Sm family of RNA-associated proteins. Molecular Biology and Evolution, 2008, 25(11): 2255-2267. DOI:10.1093/molbev/msn175 |
| [74] | VOGEL J, LUISI BF. Hfq and its constellation of RNA. Nature Reviews Microbiology, 2011, 9(8): 578-589. DOI:10.1038/nrmicro2615 |
| [75] | NIELSEN JS, BØGGILD A, ANDERSEN CBF, NIELSEN G, BOYSEN A, BRODERSEN DE, VALENTIN-HANSEN P. An Hfq-like protein in Archaea: crystal structure and functional characterization of the Sm protein from Methanococcus jannaschii. RNA: New York, N Y, 2007, 13(12): 2213-2223. DOI:10.1261/rna.689007 |
| [76] | SAUTER C, BASQUIN J, SUCK D. Sm-like proteins in eubacteria: the crystal structure of the Hfq protein from Escherichia coli. Nucleic Acids Research, 2003, 31(14): 4091-4098. DOI:10.1093/nar/gkg480 |
| [77] | HORNE M, ENGLERT C, WIMMER C, PFEIFER F. A DNA region of 9 kbp contains all genes necessary for gas vesicle synthesis in halophilic archaebacteria. Molecular Microbiology, 1991, 5(5): 1159-1174. DOI:10.1111/j.1365-2958.1991.tb01889.x |
| [78] | PFEIFER F. Haloarchaea and the formation of gas vesicles. Life: Basel, Switzerland, 2015, 5(1): 385-402. |
| [79] | ENGLERT C, HORNE M, PFEIFER F. Expression of the major gas vesicle protein gene in the halophilic archaebacterium Haloferax mediterranei is modulated by salt. Molecular and General Genetics MGG, 1990, 222(2): 225-232. |
| [80] | HECHLER T, FRECH M, PFEIFER F. Glucose inhibits the formation of gas vesicles in Haloferax volcanii transformants. Environmental Microbiology, 2008, 10(1): 20-30. |
| [81] | HECHLER T, PFEIFER F. Anaerobiosis inhibits gas vesicle formation in halophilic Archaea. Molecular Microbiology, 2009, 71(1): 132-145. DOI:10.1111/j.1365-2958.2008.06517.x |
| [82] | DASSARMA P, ZAMORA RC, MÜLLER JA, DASSARMA S. Genome-wide responses of the model archaeon Halobacterium sp. strain NRC-1 to oxygen limitation. Journal of Bacteriology, 2012, 194(20): 5530-5537. DOI:10.1128/JB.01153-12 |
| [83] | PFEIFER F. Distribution, formation and regulation of gas vesicles. Nature Reviews Microbiology, 2012, 10(10): 705-715. DOI:10.1038/nrmicro2834 |
| [84] | ENGLERT C, KRÜGER K, OFFNER S, PFEIFER F. Three different but related gene clusters encoding gas vesicles in halophilic archaea. Journal of Molecular Biology, 1992, 227(2): 586-592. DOI:10.1016/0022-2836(92)90914-6 |
| [85] | OFFNER S, WANNER G, PFEIFER F. Functional studies of the gvpACNO operon of Halobacterium salinarium reveal that the GvpC protein shapes gas vesicles. Journal of Bacteriology, 1996, 178(7): 2071-2078. DOI:10.1128/jb.178.7.2071-2078.1996 |
| [86] | RÖDER R, PFEIFER F. Influence of salt on the transcription of the gas-vesicle genes of Haloferax mediterranei and identification of the endogenous transcriptional activator gene. Microbiology: Reading, England, 1996, 142(Pt 7): 1715-1723. |
| [87] | ENGLERT C, WANNER G, PFEIFER F. Functional analysis of the gas vesicle gene cluster of the halophilic archaeon Haloferax mediterranei defines the vac-region boundary and suggests a regulatory role for the gvpD gene or its product. Molecular Microbiology, 1992, 6(23): 3543-3550. DOI:10.1111/j.1365-2958.1992.tb01789.x |
| [88] | SCHEUCH S, MARSCHAUS L, SARTORIUS-NEEF S, PFEIFER F. Regulation of gvp genes encoding gas vesicle proteins in halophilic Archaea. Archives of Microbiology, 2008, 190(3): 333-339. DOI:10.1007/s00203-008-0362-x |
| [89] | MARSCHAUS L, PFEIFER F. A dual promoter region with overlapping activator sequences drives the expression of gas vesicle protein genes in haloarchaea. Microbiology: Reading, England, 2012, 158(Pt 11): 2815-2825. |
| [90] | THOMPSON DK, PALMER JR, DANIELS CJ. Expression and heat-responsive regulation of a TFIIB homologue from the archaeon Haloferax volcanii. Molecular Microbiology, 1999, 33(5): 1081-1092. DOI:10.1046/j.1365-2958.1999.01551.x |
| [91] | LU QH, HAN J, ZHOU LG, COKER JA, DASSARMA P, DASSARMA S, XIANG H. Dissection of the regulatory mechanism of a heat-shock responsive promoter in Haloarchaea: a new paradigm for general transcription factor directed archaeal gene regulation. Nucleic Acids Research, 2008, 36(9): 3031-3042. DOI:10.1093/nar/gkn152 |
| [92] | BRÄSEN C, ESSER D, RAUCH B, SIEBERS B. Carbohydrate metabolism in Archaea: current insights into unusual enzymes and pathways and their regulation. Microbiology and Molecular Biology Reviews: MMBR, 2014, 78(1): 89-175. DOI:10.1128/MMBR.00041-13 |
| [93] | RAWLS KS, YACOVONE SK, MAUPIN-FURLOW JA. GlpR represses fructose and glucose metabolic enzymes at the level of transcription in the haloarchaeon Haloferax volcanii. Journal of Bacteriology, 2010, 192(23): 6251-6260. DOI:10.1128/JB.00827-10 |
| [94] | JOHNSEN U, SUTTER JM, SCHULZ AC, TÄSTENSEN JB, SCHÖNHEIT P. XacR-a novel transcriptional regulator of D-xylose and L-arabinose catabolism in the haloarchaeon Haloferax volcanii. Environmental Microbiology, 2015, 17(5): 1663-1676. DOI:10.1111/1462-2920.12603 |
| [95] | SUTTER JM, JOHNSEN U, SCHÖNHEIT P. Characterization of a pentonolactonase involved in D-xylose and L-arabinose catabolism in the haloarchaeon Haloferax volcanii. FEMS Microbiology Letters, 2017, 364(13): fnx140. |
| [96] | MIRFEIZOLLAHI A, YAKHCHALI B, DELDAR AA, KARKHANE AA. In silico and experimental improvement of bacteriorhodopsin production in Halobacterium salinarum R1 by increasing DNA-binding affinity of Bat through Q661R/Q665R substitutions in HTH motif. Extremophiles, 2019, 23(1): 59-67. DOI:10.1007/s00792-018-1060-5 |
| [97] | BALIGA NS, DASSARMA S. Saturation mutagenesis of the TATA box and upstream activator sequence in the haloarchaeal bop gene promoter. Journal of Bacteriology, 1999, 181(8): 2513-2518. DOI:10.1128/JB.181.8.2513-2518.1999 |
| [98] | BALIGA NS, KENNEDY SP, NG WV, HOOD L, DASSARMA S. Genomic and genetic dissection of an archaeal regulon. Proceedings of the National Academy of Sciences of the United States of America, 2001, 98(5): 2521-2525. DOI:10.1073/pnas.051632498 |
| [99] | FURTWÄNGLER K, TARASOV V, WENDE A, SCHWARZ C, OESTERHELT D. Regulation of phosphate uptake via PST transporters in Halobacterium salinarum R1. Molecular Microbiology, 2010, 76(2): 378-392. DOI:10.1111/j.1365-2958.2010.07101.x |
| [100] | STRAHL H, GREIE JC. The extremely halophilic archaeon Halobacterium salinarum R1 responds to potassium limitation by expression of the K+-transporting KdpFABC P-type ATPase and by a decrease in intracellular K+. Extremophiles, 2008, 12(6): 741-752. DOI:10.1007/s00792-008-0177-3 |
| [101] | KIXMÜLLER D, STRAHL H, WENDE A, GREIE JC. Archaeal transcriptional regulation of the prokaryotic KdpFABC complex mediating K+ uptake in H. salinarum. Extremophiles, 2011, 15(6): 643. DOI:10.1007/s00792-011-0395-y |
| [102] | KIXMÜLLER D, GREIE JC. Construction and characterization of a gradually inducible expression vector for Halobacterium salinarum, based on the kdp promoter. Applied and Environmental Microbiology, 2012, 78(7): 2100-2105. DOI:10.1128/AEM.07155-11 |
| [103] | MARTÍNEZ-ESPINOSA RM, MARHUENDA-EGEA FC, BONETE MJ. Assimilatory nitrate reductase from the haloarchaeon Haloferax mediterranei: purification and characterisation. FEMS Microbiology Letters, 2001, 204(2): 381-385. DOI:10.1111/j.1574-6968.2001.tb10914.x |
| [104] | PASTOR-SOLER S, CAMACHO M, BAUTISTA V, BONETE MJ, ESCLAPEZ J. Towards the elucidation of assimilative nasABC operon transcriptional regulation in Haloferax mediterranei. Genes, 2021, 12(5): 619. DOI:10.3390/genes12050619 |
| [105] | WALSBY AE. Gas vesicles. Microbiological Reviews, 1994, 58(1): 94-144. DOI:10.1128/mr.58.1.94-144.1994 |
| [106] | MAGER WH, de KRUIJFF AJ. Stress-induced transcriptional activation. Microbiological Reviews, 1995, 59(3): 506-531. DOI:10.1128/mr.59.3.506-531.1995 |
| [107] | ZUBER U, SCHUMANN W. CIRCE, a novel heat shock element involved in regulation of heat shock operon dnaK of Bacillus subtilis. Journal of Bacteriology, 1994, 176(5): 1359-1363. DOI:10.1128/jb.176.5.1359-1363.1994 |
| [108] | SORGER PK. Heat shock factor and the heat shock response. Cell, 1991, 65(3): 363-366. DOI:10.1016/0092-8674(91)90452-5 |
| [109] | THOMPSON DK, DANIELS CJ. Heat shock inducibility of an archaeal TATA-like promoter is controlled by adjacent sequence elements. Molecular Microbiology, 1998, 27(3): 541-551. DOI:10.1046/j.1365-2958.1998.00698.x |
| [110] | KARAN R, CAPES MD, DASSARMA P, DASSARMA S. Cloning, overexpression, purification, and characterization of a polyextremophilic β-galactosidase from the Antarctic haloarchaeon Halorubrum lacusprofundi. BMC Biotechnology, 2013, 13: 3. DOI:10.1186/1472-6750-13-3 |
| [111] | MARTIN JH, SHERWOOD RAWLS K, CHAN JC, HWANG S, MARTINEZ-PASTOR M, MCMILLAN LJ, PRUNETTI L, SCHMID AK, MAUPIN-FURLOW JA. GlpR is a direct transcriptional repressor of fructose metabolic genes in Haloferax volcanii. Journal of Bacteriology, 2018, 200(17): e00244-e00218. |
| [112] | GROPP F, BETLACH MC. The bat gene of Halobacterium halobium encodes a trans-acting oxygen inducibility factor. Proceedings of the National Academy of Sciences of the United States of America, 1994, 91(12): 5475-5479. DOI:10.1073/pnas.91.12.5475 |
| [113] | VERSHININA OA, ZNAMENSKAIA LV. Pho regulony bakteriĭ. Mikrobiologiia, 2002, 71(5): 581-595. |
| [114] | LAM WL, COHEN A, TSOULUHAS D, DOOLITTLE WF. Genes for tryptophan biosynthesis in the archaebacterium Haloferax volcanii. Proceedings of the National Academy of Sciences of the United States of America, 1990, 87(17): 6614-6618. DOI:10.1073/pnas.87.17.6614 |
| [115] | LAM WL, LOGAN SM, DOOLITTLE WF. Genes for tryptophan biosynthesis in the halophilic archaebacterium Haloferax volcanii: the trpDFEG cluster. Journal of Bacteriology, 1992, 174(5): 1694-1697. DOI:10.1128/jb.174.5.1694-1697.1992 |
| [116] | MALKI L, YANKU M, BOROVOK I, COHEN G, MEVARECH M, Aharonowitz Y. Identification and characterization of gshA, a gene encoding the glutamate-cysteine ligase in the halophilic archaeon Haloferax volcanii. Journal of Bacteriology, 2009, 191(16): 5196-5204. DOI:10.1128/JB.00297-09 |
| [117] |
YU HM, ZHENG YK, DU Y, WANG MM, LIANG YX. Microbial promoter engineering strategies in synthetic biology. Synthetic Biology Journal, 2021, 2(4): 598-611.
(in Chinese) 于慧敏, 郑煜堃, 杜岩, 王苗苗, 梁有向. 合成生物学研究中的微生物启动子工程策略. 合成生物学, 2021, 2(4): 598-611. |
| [118] | BORN J, WEITZEL K, SUESS B, PFEIFER F. A synthetic riboswitch to regulate haloarchaeal gene expression. Frontiers in Microbiology, 2021, 12: 696181. DOI:10.3389/fmicb.2021.696181 |
 2023, Vol. 63
2023, Vol. 63