
中国科学院微生物研究所,中国微生物学会
文章信息
- 翟康乐, 宫雅楠, 肖迪, 孙路, 何利华, 房梦飏, 尤元海, 王磊, 闫笑梅, 张建中. 2023
- ZHAI Kangle, GONG Yanan, XIAO Di, SUN Lu, HE Lihua, FANG Mengyang, YOU Yuanhai, WANG Lei, YAN Xiaomei, ZHANG Jianzhong.
- 幽门螺杆菌分子伴侣GroEL相互作用蛋白分析
- Interacting proteins of Helicobacter pylori GroEL protein
- 微生物学报, 63(5): 1970-1981
- Acta Microbiologica Sinica, 63(5): 1970-1981
-
文章历史
- 收稿日期:2023-02-27
- 网络出版日期:2023-04-23
2. 南京医科大学公共卫生学院, 江苏 南京 211166
2. School of Public Health, Nanjing Medical University, Nanjing 211166, Jiangsu, China
幽门螺杆菌(Helicobacter pylori, HP)是革兰染色阴性、呈螺旋状的微需氧菌。HP持续感染可引发胃部慢性活动性炎症[1],其中约15%‒20%会发生消化性溃疡[2],5%‒10%发生相关性消化不良[3],约1%会发展为胃恶性肿瘤(胃癌、黏膜相关淋巴组织淋巴瘤)[4]。HP致病作用广泛,疾病负担重,但致病机制尚未完全明确。
HP的分子伴侣GroEL,是热休克蛋白60 (heat shock protein, HSP60)在原核生物中的同源蛋白,其主要功能是帮助蛋白转运、完成正确折叠及恢复天然构象[5]。在HP相关研究中发现,GroEL也是一种重要的致病相关抗原成分,超过80%的HP感染人群可产生针对GroEL的抗体[6-7],且GroEL抗体的持续存在被认为与HP引起的胃部病变进展为胃癌存在关联[8-9],GroEL也可能在HP诱发炎症向胃癌进展过程中发挥重要作用。由于HP的GroEL与人HSP60同源性较高,其抗体介导的交叉反应在动脉粥样硬化的发生中起到一定的作用[10]。作为一种重要的分子伴侣毒力因子,GroEL在HP生物学中的作用非常广泛和重要,虽然人类对HP的分子生物学研究已经有40年的历史,但目前仍缺乏对GroEL与其他蛋白质相互作用的系统性研究。本研究拟通过表达、纯化获得带有His标签的GroEL蛋白,用免疫沉淀法捕获HP中可能与GroEL结合的蛋白质,通过对可能与GroEL结合的蛋白质组成谱的解析和相互关系网络分析,为GroEL及其相关蛋白在HP致病中的作用机制研究及可能的感染防控靶点探索提供新的思路。
1 材料与方法 1.1 菌株、质粒及试剂HP标准菌株ATCC 43504购自美国典型微生物菌种保藏中心(American Type Culture Collection, ATCC)并由本实验室保藏,感受态细胞E. coli DH5α、E. coli BL21(DE3)、表达质粒pET-28a(+)及5K DNA Marker购自北京全式金生物技术股份有限公司,基因组提取试剂盒QIAamp DNA Mini Kit购自QIAGEN公司,限制性内切酶BamH I、Xho I和Q5®超保真2×PCR预混液购自纽英伦生物技术(北京)有限公司,质粒提取试剂盒E.Z.N.A.® Plasmid DNA Mini Kit、胶回收试剂盒E.Z.N.A.® Gel Extraction Kit购自OMEGA公司,T4 DNA连接酶购自TaKaRa公司,His标签蛋白纯化磁珠HisPurTM Ni-NTA、免疫沉淀磁珠DynabeadsTM Protein G Immunoprecipitation Kit购自赛默飞世尔科技(中国)有限公司,鼠抗His标签单抗和鼠抗GST标签单抗购自艾博抗(上海)贸易有限公司,蛋白marker和咪唑(分析纯)购自北京索莱宝科技有限公司,培养基及蛋白纯化、质谱鉴定所用缓冲液等其他试剂均按照需要购买或配制。
1.2 引物设计和合成从NCBI获取ATCC 43504基因组数据(GCF_900478294.1),提取groEL基因序列并设计引物,引物序列为groEL-F:5′-CGGGATCC (BamH Ⅰ) ATGGCAAAAGAAATCAAATTTTCA G-3′;groEL-R:5′-CCGCTCGAG (Xho Ⅰ) CATC ATTCCGCCCATGCCT-3′。引物由北京奥科鼎盛生物科技有限公司合成。
1.3 groEL基因PCR扩增刮取培养48 h的新鲜ATCC 43504菌体,提取其基因组DNA用作模板,分别以groEL-F和groEL-R为上、下游引物,进行PCR扩增。25 μL扩增体系:模板(150 ng/μL) 1 μL,上、下游引物各(10 μmol/L) 0.5 μL,2×PCR预混液12.5 μL,ddH2O 10.5 μL。扩增程序:94 ℃ 5 min;94 ℃ 30 s,65 ℃ 30 s,72 ℃ 90 s,30个循环;72 ℃ 10 min,4 ℃保存。采用1.5%琼脂糖凝胶电泳进行PCR产物检测,胶回收试剂盒纯化PCR产物。
1.4 groEL原核表达系统构建用BamH I和Xho I分别酶切1 μg的pET-28a(+)质粒和1 μg的胶回收纯化groEL,经1.5%琼脂糖凝胶电泳后切胶回收,测量浓度后将groEL片段和线性化的pET-28a(+)按3:1比例混合,加入T4 DNA连接酶16 ℃过夜,连接产物热激转化E. coli DH5α,挑取阳性单克隆菌落,37 ℃过夜培养,经菌液PCR鉴定groEL基因阳性后送生工生物工程(上海)股份有限公司进行测序确认。选择测序结果正确的E. coli DH5α克隆,提取重组表达质粒pET-28a(+)-groEL后热激转化E. coli BL21(DE3),挑选阳性单克隆,培养过夜,经菌液PCR鉴定groEL基因阳性后,即完成原核表达重组菌E. coli BL21(DE3)pET-28a(+)-groEL的构建。
1.5 GroEL诱导表达将E. coli BL21(DE3)pET-28a(+)-groEL置于LB液体培养基中,37 ℃振荡培养至OD600为0.6‒1.0时,加入1.0 mmol/L IPTG,37 ℃、180 r/min振荡培养4 h,诱导GroEL蛋白表达。离心收集菌体,用磷酸盐缓冲溶液(phosphate buffer saline, PBS)重悬,冰浴超声裂解,高速离心,分别收集上清和沉淀,SDS-PAGE检测目的蛋白的表达情况。
1.6 GroEL纯化及鉴定将500 mL诱导后的菌液离心后,用25 mL含50 mmol咪唑的平衡缓冲液重悬菌体,冰浴超声裂解,高速离心收集上清,即为待纯化样品,按400 μL待纯化样品用40 μL HisPurTM Ni-NTA磁珠的比例进行纯化。将待纯化样品加入磁珠中,涡旋10 s,孵育30 min,用含100 mmol/L咪唑的洗涤缓冲液洗涤磁珠2次,用含300 mmol/L咪唑的洗脱缓冲液洗脱2次,分别收集洗脱液,采用二辛可宁酸(bicinchonininc acid, BCA)法测定洗脱液中蛋白浓度并经SDS-PAGE检测纯化效果。40 μL含GroEL的洗脱液经50 mmol/L碳酸氢铵(NH4HCO3) 3次超滤换液后,加入1 μL 200 mmol/L三(2-羧乙基)膦[tris(2-carboxyethyl)phosphine, TCEP]室温静置1 h,加入1 μL 375 mmol/L碘乙酰胺(iodoacetamide, IAA)室温避光静置30 min,加入1 μg胰酶37 ℃过夜,用Pierce® C18 Spin Columns脱盐柱脱盐后,4 ℃真空离心浓缩,用10 μL 0.1%甲酸(formic acid, FA)重悬后,经高效液相色谱-串联质谱(HPLC-MS/MS)获取原始谱图。用Proteome Discoverer (V. 2.4)软件对原始谱图进行蛋白库(UniProtKB/Swiss-Prot)搜索,分析鉴定相应肽段及所匹配到的蛋白质。
1.7 HP ATCC 43504中可能和GroEL结合的蛋白质的鉴定及分类将2 μg鼠抗His单抗与50 μL (1.5 mg) Protein G磁珠室温孵育10 min,结合至Protein G磁珠上;将新鲜培养的ATCC 43504用PBS洗涤2次,离心收集菌体,按1 g湿重菌体/10 mL PBS的比例重悬,冰浴超声裂解,高速离心后收集上清,即为ATCC 43504蛋白提取液。向500 μL ATCC 43504蛋白提取液中加入5 μg纯化后的GroEL蛋白,室温孵育1 h,加至结合了鼠抗His单抗的Protein G磁珠中,室温孵育10 min,用200 μL洗涤液洗涤磁珠3次,加入20 μL洗脱液振荡孵育2 min,收集洗脱液用于下游质谱鉴定。将2 μg鼠抗GST单抗结合至50 μL (1.5 mg) Protein G磁珠上,用作阴性对照,其他条件与实验组一致。洗脱液中蛋白质的质谱鉴定方法同1.6所述。在Uniprot蛋白数据库(https://www.uniprot.org/)中查阅鉴定出的蛋白质信息,并根据主要功能进行分类。
1.8 GroEL及可能和其结合的蛋白质的相互关系网络分析在STRING (search tool for the retrival of interacting genes/proteins)数据库(https://string-db.org/, version 11.5)中,首先选择“Protein by name”功能,“Protein name”输入”GroEL”,“Organisms”选择“Helicobacter pylori 26695”,查询数据库中预测的GroEL功能伙伴蛋白,然后选择“Multiple proteins”功能,“List of names”中导入实验中鉴定出的蛋白质列表,“Organisms”选择“Helicobacter pylori 26695”,进行多个蛋白质的相互关系网络分析。
2 结果与分析 2.1 groEL基因PCR扩增以ATCC 43504基因组DNA为模板进行PCR扩增,1.5%琼脂糖凝胶电泳显示扩增产物条带介于1 500–2 000 bp之间(图 1),产物大小符合预期(1 655 bp)。
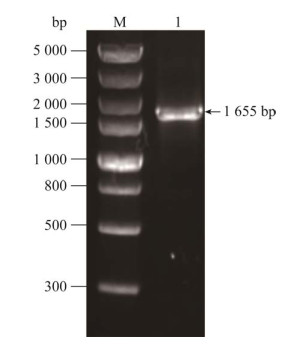
|
| 图 1 groEL基因PCR扩增结果 Figure 1 Amplification results of groEL gene by PCR. M: DNA marker; 1: PCR amplification product of the groEL gene. |
2.2 GroEL表达系统构建、表达和纯化
经菌液PCR鉴定groEL基因阳性,并通过测序确认目的片段与参考序列完全一致,成功构建了原核表达重组菌并命名为E. coli BL21(DE3)pET-28a(+)-groEL。通过加入1.0 mmol/L IPTG,在37 ℃、180 r/min振荡培养的诱导条件下,SDS-PAGE显示目的蛋白条带介于63–75 kDa,大小符合预期,目的条带在沉淀中占较大比重,上清中也有一定表达(图 2),表明目的蛋白在诱导过程中能大量表达,表达后的目的蛋白能以可溶性形式存在,但主要形成包涵体。可溶性蛋白经Ni-NTA磁珠亲和纯化后,SDS-PAGE显示洗脱液中目的蛋白条带明显,且纯度较高(图 3)。
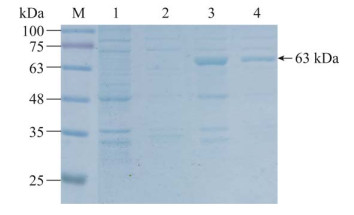
|
| 图 2 GroEL蛋白的诱导表达结果 Figure 2 Induced expression results of GroEL protein. M: Protein marker; 1: Precipitation of the cell lysate from uninduced E. coli BL21(DE3)pET-28a(+)-groEL cells; 2: Supernatant of the cell lysate from uninduced E. coli BL21(DE3)pET-28a(+)-groEL cells; 3: Precipitation of the cell lysate from induced E. coli BL21(DE3)pET-28a(+)-groEL cells; 4: Supernatant of the cell lysate from induced E. coli BL21(DE3)pET-28a(+)-groEL cells. |
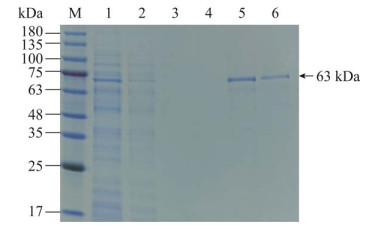
|
| 图 3 GroEL蛋白纯化结果 Figure 3 Purification results of GroEL protein. M: Protein marker; 1: Sample before Ni-NTA magnetic beads incubation; 2: Sample after Ni-NTA magnetic beads incubation; 3, 4: The first and second magnetic beads washing solution; 5, 6: The first and second magnetic beads elution solution. |
2.3 GroEL蛋白的质谱鉴定
纯化后的目的蛋白经质谱鉴定,结果显示其与HP的GroEL蛋白相匹配并且获得极高的质谱评分,表明原核表达重组菌E. coli BL21(DE3)pET-28a(+)-groEL表达目的蛋白GroEL成功。
2.4 HP ATCC 43504中可能和GroEL结合的蛋白鉴定及分类从ATCC 43504的蛋白提取液中捕获了可能与GroEL结合的蛋白质,经质谱鉴定出59种蛋白,具体信息见表 1。对蛋白做功能分类发现,主要包括19种代谢酶类(参与氧化还原相关酶类7种,包括KatA、GltA、AhpC等;肽酶5种,PepA、RocF、HtrA等;参与脂肪代谢酶类2种;参与ATP合成酶类2种;尿素酶2种)、15种外膜蛋白(黏附素BabA、SabA、HapA及其他膜蛋白等)、8种转录翻译相关蛋白(Tuf、RpoBC等)、5种分子伴侣(DnaK、GroES等)、3种细胞毒素相关蛋白(CagA等)、3种氧化还原相关蛋白(TrxA等)、2种信号转导蛋白(TlpB、TlpD)和4种功能不明蛋白,具体如表 2所示。
| Accession No. | Description | Coverage (%) | Peptides | PSMs | Unique peptides | AAs | MW (kDa) | calc pI | Score |
| WP_000961643.1 | Peroxiredoxin (AhpC) | 35 | 6 | 9 | 6 | 198 | 22.2 | 6.30 | 25.54 |
| WP_108169803.1 | Hop family adhesin HopQ (HopQ) | 17 | 7 | 8 | 7 | 641 | 69.7 | 9.11 | 21.25 |
| WP_108169269.1 | Hop family adhesin BabA (BabA) | 13 | 8 | 8 | 8 | 744 | 80.4 | 8.81 | 20.33 |
| WP_001040578.1 | Elongation factor Tu (Tuf) | 33 | 9 | 9 | 9 | 399 | 43.6 | 5.26 | 20.19 |
| WP_108169606.1 | Methyl-accepting chemotaxis protein TlpB (TlpB) | 17 | 8 | 8 | 8 | 565 | 62.7 | 6.42 | 17.21 |
| WP_000724287.1 | Urease subunit beta (UreB) | 12 | 6 | 6 | 6 | 569 | 61.7 | 6.01 | 16.52 |
| WP_108169537.1 | Outer membrane beta-barrel protein | 9 | 5 | 6 | 5 | 699 | 76.0 | 8.53 | 16.00 |
| WP_108169053.1 | Hop family adhesin AlpA (AlpA) | 14 | 4 | 5 | 4 | 518 | 56.1 | 9.10 | 14.20 |
| WP_000795968.1 | LPP20 family lipoprotein (Lpp20) | 26 | 5 | 5 | 5 | 175 | 19.1 | 9.51 | 12.97 |
| WP_108169033.1 | Catalase (KatA) | 10 | 4 | 5 | 4 | 505 | 58.6 | 8.66 | 12.29 |
| WP_000323696.1 | DUF3944 domain-containing protein (NY40_0613) | 24 | 5 | 5 | 5 | 253 | 28.4 | 5.43 | 12.24 |
| WP_000779223.1 | Urease subunit alpha (UreA) | 13 | 4 | 5 | 4 | 238 | 26.5 | 8.47 | 12.21 |
| WP_108169170.1 | Outer membrane beta-barrel protein HofC (B0X38_04460) | 9 | 4 | 5 | 4 | 528 | 59.4 | 9.33 | 12.09 |
| WP_108168926.1 | Sialic acid-binding protein (SabA) | 32 | 4 | 5 | 4 | 152 | 17.6 | 8.72 | 11.67 |
| WP_000080506.1 | F0F1 ATP synthase subunit alpha (AtpA) | 9 | 4 | 4 | 4 | 503 | 55.1 | 5.40 | 9.41 |
| WP_001861912.1 | F0F1 ATP synthase subunit beta (AtpD) | 10 | 4 | 4 | 4 | 466 | 51.1 | 5.34 | 9.00 |
| WP_000671927.1 | Co-chaperone GroES (GroES) | 23 | 3 | 3 | 3 | 118 | 13.0 | 6.60 | 8.92 |
| WP_108169412.1 | TonB-dependent receptor (FrpB) | 6 | 4 | 4 | 4 | 863 | 95.9 | 9.14 | 8.36 |
| WP_108169054.1 | Hop family adhesin AlpB (AlpB) | 7 | 3 | 3 | 3 | 528 | 56.8 | 9.19 | 8.32 |
| WP_108169330.1 | Hop family outer membrane protein HopA (HopA) | 10 | 3 | 3 | 3 | 482 | 52.8 | 9.44 | 7.23 |
| WP_108168983.1 | MULTISPECIES: flagellar sheath lipoprotein HpaA (Proteobacteria) (HpaA) | 16 | 3 | 3 | 3 | 260 | 29.1 | 8.27 | 7.02 |
| WP_097551802.1 | Tumor necrosis factor alpha-inducing protein (TipA) | 21 | 3 | 3 | 3 | 192 | 21.8 | 8.51 | 6.96 |
| WP_108169924.1 | Chemotaxis chemoreceptor TlpD (TlpD) | 9 | 3 | 3 | 3 | 433 | 48.4 | 6.25 | 6.84 |
| WP_108169893.1 | Type IV secretion system oncogenic effector CagA (CagA) | 3 | 3 | 3 | 3 | 1 247 | 139.0 | 8.84 | 6.66 |
| WP_162296893.1 | S41 family peptidase (C2R77_03000) | 6 | 3 | 3 | 3 | 454 | 50.0 | 9.38 | 6.56 |
| WP_108169905.1 | Leucyl aminopeptidase (PepA) | 8 | 3 | 3 | 3 | 496 | 54.4 | 7.09 | 6.11 |
| WP_033765600.1 | Outer membrane protein Omp18 (Omp18) | 18 | 2 | 2 | 2 | 179 | 19.9 | 6.09 | 5.57 |
| WP_108169611.1 | Molecular chaperone DnaK (DnaK) | 4 | 2 | 2 | 2 | 620 | 67.1 | 5.10 | 5.49 |
| WP_108169113.1 | HpaA2 protein (HpaA2) | 10 | 2 | 2 | 2 | 249 | 28.3 | 7.39 | 5.38 |
| WP_108169113.1 | 30S ribosomal protein S12 (RpsL) | 10 | 2 | 2 | 2 | 249 | 28.3 | 7.39 | 5.38 |
| WP_000020199.1 | Thioredoxin (TrxA) | 25 | 2 | 2 | 2 | 106 | 11.8 | 5.22 | 5.01 |
| WP_000086604.1 | 50S ribosomal protein L6 (RplF) | 10 | 2 | 2 | 2 | 178 | 19.5 | 9.83 | 4.72 |
| WP_108169722.1 | 50S ribosomal protein L2 (RplB) | 8 | 2 | 2 | 2 | 276 | 30.2 | 10.36 | 4.50 |
| WP_001018219.1 | 50S ribosomal protein L7/L12 (RplL) | 21 | 2 | 2 | 2 | 125 | 13.3 | 4.98 | 4.41 |
| WP_162296871.1 | Bifunctional aconitate hydratase 2/2-methylisocitrate dehydratase (HP17_08079) | 3 | 2 | 2 | 2 | 852 | 92.6 | 6.57 | 4.38 |
| WP_108169000.1 | Thioredoxin-disulfide reductase (TrxB) | 8 | 2 | 2 | 2 | 311 | 33.4 | 6.09 | 4.32 |
| WP_108169788.1 | Elongation factor G (FusA) | 2 | 2 | 2 | 2 | 692 | 77.0 | 5.39 | 4.15 |
| WP_108169306.1 | AAA family ATPase (EC537_00185) | 3 | 2 | 2 | 2 | 856 | 96.5 | 6.13 | 4.08 |
| WP_108168981.1 | Trigger factor (Tig) | 4 | 2 | 2 | 2 | 451 | 51.8 | 5.31 | 3.89 |
| WP_108169883.1 | Cag pathogenicity island protein Cag1 (BHU51_05410) | 12 | 1 | 1 | 1 | 115 | 12.4 | 8.44 | 2.71 |
| WP_000889333.1 | 50S ribosomal protein L25/general stress protein Ctc (RplY) | 5 | 1 | 1 | 1 | 178 | 19.9 | 9.39 | 2.64 |
| WP_108169809.1 | Outer membrane beta-barrel protein HofH (AOD76_0206895) | 3 | 1 | 1 | 1 | 471 | 52.7 | 9.48 | 2.36 |
| WP_108169980.1 | Zinc ribbon domain-containing protein | 2 | 1 | 1 | 1 | 412 | 43.6 | 6.65 | 2.22 |
| WP_108169980.1 | Beta-ketoacyl-ACP synthase II (BB414_08470) | 2 | 1 | 1 | 1 | 412 | 43.6 | 6.65 | 2.22 |
| WP_108169314.1 | Hop family outer membrane protein HopF (HopF) | 1 | 1 | 1 | 1 | 485 | 53.1 | 9.41 | 2.17 |
| WP_000117433.1 | Citrate synthase (GltA) | 3 | 1 | 1 | 1 | 426 | 48.3 | 7.88 | 2.09 |
| WP_001183652.1 | Fumarate reductase cytochrome b subunit (FrdC) | 4 | 1 | 1 | 1 | 255 | 28.8 | 9.44 | 2.04 |
| WP_108169352.1 | Acyl-ACP-UDP-N-acetylglucosamine O-acyltransferase (LpxA) | 4 | 1 | 1 | 1 | 270 | 29.8 | 7.08 | 2.04 |
| WP_108169612.1 | Nucleotide exchange factor GrpE (GrpE) | 7 | 1 | 1 | 1 | 191 | 22.2 | 5.41 | 2.02 |
| WP_108169634.1 | Cytochrome-c oxidase, cbb3-type subunit III (OUK_0478) | 3 | 1 | 1 | 1 | 292 | 32.6 | 6.18 | 2.02 |
| WP_000976634.1 | Do family serine endopeptidase | 1 | 1 | 1 | 1 | 476 | 51.5 | 9.19 | 2.02 |
| WP_108169978.1 | Cag pathogenicity island type IV secretion system protein CagD (CagD) | 4 | 1 | 1 | 1 | 207 | 24.0 | 8.16 | 1.99 |
| WP_108169513.1 | ABC transporter substrate-binding protein | 4 | 1 | 1 | 1 | 333 | 37.2 | 8.95 | 1.97 |
| WP_108169787.1 | DNA-directed RNA polymerase subunit beta/beta′ (RpoBC) | 0 | 1 | 1 | 1 | 2 890 | 323.3 | 7.06 | 1.95 |
| WP_108169488.1 | arginase (RocF) | 4 | 1 | 1 | 1 | 322 | 36.8 | 6.99 | 1.93 |
| WP_108168925.1 | DUF1104 domain-containing protein (AA977_03345) | 10 | 1 | 1 | 1 | 140 | 16.0 | 9.52 | 1.92 |
| WP_108169590.1 | FAD-dependent oxidoreductase (Mqo) | 2 | 1 | 1 | 1 | 450 | 50.7 | 7.64 | 1.92 |
| WP_108169497.1 | Cytochrome c1 (HPK25_00925) | 3 | 1 | 1 | 1 | 285 | 31.7 | 8.54 | 1.92 |
| WP_108169917.1 | 2-oxoglutarate synthase subunit alpha (BZK19_00840) | 2 | 1 | 1 | 1 | 375 | 41.4 | 6.40 | 1.90 |
| AA: Length of protein; calc: pI, calculated isoelectric point; MW: Molecular weight; PSM: Peptide spectrum match. | |||||||||
| Protein group | Proteins | Count* |
| Enzyme | 19 | |
| Redox-related enzyme | GltA, KatA, AhpC, TrxB, FrdC, Mqo, BZK19_00840 | 7 |
| Peptidase | PepA, RocF, HtrA, AA977_03345, C2R77_03000 | 5 |
| Other enzymes | UreA, UreB, AtpA, AtpD, LpxA, BB414_08470, HP17_08079 | 7 |
| Outer membrane protein | FrpB, AlpA, AlpB, AOD76_0206895, B0X38_04460, BabA, HopA, HopF, HopQ, HpaA, HpaA2, Omp18, SabA, Outer membrane beta-barrel protein, Lpp20 | 15 |
| Transcription-translation-associated proteins | FusA, RplB, RplF, RplL, RplY, RpoBC, RpsL, Tuf | 8 |
| Chaperone | DnaK, GroES, GrpE, Tig, ClpB | 5 |
| Cag pathogenicity island related proteins | CagA, CagD, BHU51_05410 | 3 |
| Redox-related proteins | TrxA, HPK25_00925, OUK_0478 | 3 |
| Signal transduction related proteins | TlpB, TlpD | 2 |
| Proteins with function not established | AA977_03345, TipA, Zinc ribbon domain-containing protein, ABC transporter substrate-binding protein | 4 |
| *: Indicates the number of proteins classified into the group. | ||
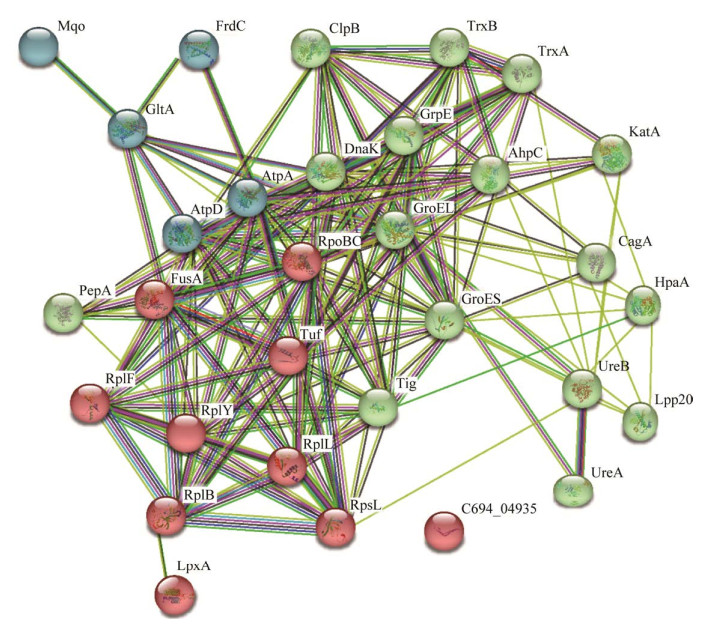
2.5 GroEL和其可能结合蛋白的相互关系网络分析
STRING数据库中预测的GroEL功能伙伴蛋白有10种,其中7种为分子伴侣,另外3种为柠檬酸合酶(GltA)、F0F1 ATP合酶亚基β (AptD)和DNA导向的RNA聚合酶亚基β/β (RpoBC) (图 4)。本研究鉴定出的59种蛋白,30种在STRING数据库中有收录,其中8种是数据库中预测的GroEL功能伙伴蛋白。STRING数据库分别从基因邻接、基因融合、基因共现、基因共表达、实验数据、数据库数据、文本挖掘数据和同源性8个方面对蛋白的相互关系进行评分,高评分表示蛋白之间发生相互作用的可能性越大,本研究中鉴定出的蛋白质与GroEL的关系紧密度评分详见表 3。相互关系网络分析发现GroEL处于中心节点,整个网络可形成3个较为明显的分簇,如图 5所示,最大的簇是以绿色节点表示的16种蛋白所形成的网络,主要包括分子伴侣、尿素酶(UreA、UreB)、硫氧还蛋白(TrxA)、硫氧还蛋白二硫化物还原酶(TrxB)、过氧化氢酶(KatA)、黏附素HpaA和细胞毒素相关蛋白CagA等;其次是以红色节点表示的10种蛋白所形成的网络,主要是转录和翻译相关的蛋白;最小的簇是以蓝色节点表示的5种蛋白所形成的网络,包括3种与三羧酸循环相关的酶(GltA、FrdC、Mqo)和2种与ATP合成相关的酶(AtpA、AtpD)。
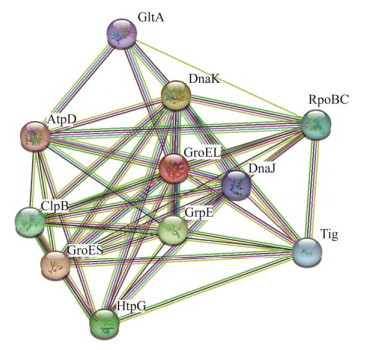
|
| 图 4 GroEL与其预测的功能伙伴相互关系的网络分析 Figure 4 Network analysis of the relationship between GroEL and its predicted functional partners. |
| Protein | Neighborhood | Gene fusion | Cooccurence | Homology | Coexpression | Experiments | Databases | Textmining | Score |
| GroES | 0.832 | 0 | 0.438 | 0 | 0.874 | 0.957 | 0 | 0.945 | 0.999 |
| DnaK | 0.081 | 0 | 0.462 | 0 | 0.871 | 0.335 | 0 | 0.933 | 0.996 |
| GrpE | 0.163 | 0 | 0.212 | 0 | 0.869 | 0.133 | 0.449 | 0.894 | 0.994 |
| ClpB | 0.060 | 0 | 0 | 0 | 0.870 | 0 | 0 | 0.889 | 0.985 |
| RpoBC | 0 | 0 | 0 | 0 | 0.086 | 0.959 | 0 | 0.611 | 0.984 |
| Tig | 0 | 0 | 0 | 0 | 0.096 | 0.300 | 0 | 0.882 | 0.918 |
| GltA | 0 | 0 | 0 | 0 | 0.071 | 0.074 | 0.572 | 0.791 | 0.913 |
| AtpD | 0.070 | 0 | 0 | 0 | 0.062 | 0.043 | 0.565 | 0.794 | 0.911 |
| AhpC | 0 | 0 | 0 | 0 | 0.234 | 0.139 | 0.138 | 0.827 | 0.889 |
| AtpA | 0 | 0 | 0 | 0 | 0.089 | 0.094 | 0.572 | 0.678 | 0.871 |
| FusA | 0 | 0 | 0 | 0 | 0.146 | 0.297 | 0 | 0.793 | 0.865 |
| RplL | 0 | 0 | 0.245 | 0 | 0.093 | 0 | 0 | 0.808 | 0.857 |
| Tuf | 0 | 0 | 0 | 0 | 0.265 | 0.138 | 0 | 0.793 | 0.857 |
| UreA | 0.117 | 0 | 0 | 0 | 0 | 0.788 | 0 | 0.281 | 0.853 |
| UreB | 0.043 | 0 | 0 | 0 | 0 | 0.319 | 0 | 0.768 | 0.835 |
| RplY | 0 | 0 | 0 | 0 | 0.081 | 0 | 0 | 0.791 | 0.800 |
| KatA | 0 | 0 | 0 | 0 | 0.061 | 0 | 0 | 0.794 | 0.798 |
| CagA | 0 | 0 | 0 | 0 | 0.058 | 0 | 0 | 0.767 | 0.771 |
| HpaA | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.769 | 0.769 |
| TrxA | 0.043 | 0 | 0 | 0 | 0.072 | 0.338 | 0 | 0.645 | 0.763 |
| TrxB | 0.043 | 0 | 0 | 0 | 0.065 | 0 | 0 | 0.696 | 0.704 |
| PepA | 0.071 | 0 | 0 | 0 | 0.117 | 0.134 | 0 | 0.506 | 0.602 |
| Lpp20 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.542 | 0.542 |
| RplB | 0 | 0 | 0 | 0 | 0.156 | 0 | 0 | 0.376 | 0.450 |
| RpsL | 0 | 0 | 0 | 0 | 0.150 | 0 | 0 | 0.363 | 0.435 |
|
| 图 5 GroEL与其可能结合蛋白相互关系的网络分析 Figure 5 Network analysis of the relationship between GroEL and its possible binding proteins. |
3 讨论与结论
在进行免疫沉淀或者Pulldown实验时,降低本底非特异性结合非常重要。本研究选择Protein G磁珠结合抗His标签抗体免疫沉淀的方法,可以从以下几个方面降低非特异性结合。首先,利用纯化后的GroEL蛋白与HP全菌蛋白提取液进行相互作用,可最大限度减少可能存在的高丰度E. coli蛋白的影响;其次,经过抗His标签抗体预先与Protein G磁珠结合,可以起到封闭磁珠及Protein G的作用,降低杂蛋白与磁珠及Protein G的非特异性结合。因此,当实验组中加入的GroEL蛋白含量占绝对优势时,极少量的E. coli杂蛋白及HP蛋白提取液中的非特异性结合蛋白对实验结果的影响是较为有限的。
作为分子伴侣,GroEL通常在细胞质中形成具有2个空腔的圆柱形寡聚体,并在GroES的协助下完成对底物蛋白的重新折叠。在大肠杆菌中,GroEL只能折叠其蛋白质组10%的蛋白[11],说明其折叠的底物存在一定的偏好性。进一步的研究发现,大肠杆菌中GroEL结合底物中丰度最高的52种蛋白主要为氨基酸、糖等代谢相关的酶类及参与转录翻译的蛋白质[12]。本研究较为系统地鉴定出了HP中可能与GroEL结合的59种蛋白质,初步揭示了HP中分子伴侣GroEL的底物蛋白种类和数量,这为深入了解GroEL的生物学功能提供了重要的参考。
本研究鉴定出的蛋白质中,有19种参与各种代谢的酶类,8种参与转录翻译的蛋白,表明GroEL在维持代谢和蛋白质转录翻译中扮演着重要角色。本研究发现15种外膜蛋白,包括BabA、HpaA、SabA等重要黏附素,可能与GroEL结合。先前的研究表明,GroEL可定位在HP外膜上[13],也可出现在HP产生的膜外囊泡中[14],这都提示GroEL可能与外膜蛋白存在广泛的关联,但目前GroEL通过何种途径从胞质转位到外膜,以及其在外膜和膜外囊泡中的作用尚不明确。有研究报道,在斑疹伤寒立克次体中,外膜蛋白OmpB与GroEL在胞质内结合后共同转位到外膜[15],对于HP中的GroEL,是否也是外膜蛋白与之结合后共转位到外膜上,需要进一步研究证实。此外,有研究报道,GroEL与外膜蛋白OMPc是支气管败血博德特氏菌(Bordetella septicaemia)来源的膜外囊泡中的主要抗原,可以诱导小鼠产生保护性免疫反应[16],HP膜外囊泡中的GroEL能否发挥类似的作用,值得进一步探索。本研究发现,有10种与氧化还原相关的酶和蛋白可能与GroEL结合,提示GroEL在HP面临氧化应激时对于维持这些酶和蛋白的活性可能起作用,已有研究报道GroEL可协同亚砜还原酶(sulfoxide reductase)修复因氧化受损的过氧化氢酶(KatA)并恢复其活性[17]。本研究发现,维持HP生存至关重要的尿素酶(UreA和UreB亚单位)可能与GroEL结合,提示GroEL在帮助HP适应严苛的胃内环境方面发挥着作用,其中UreA与GroEL的相互作用已经得到实验证实[18-19]。此外,HP最重要的毒力因子CagA也可能与GroEL结合,提示GroEL可能参与CagA引起的机体病理损伤过程。
综上所述,本研究报道了HP中可与GroEL相互作用的蛋白质组成谱,功能分类和相互关系网络分析发现GroEL结合的蛋白质种类繁多,功能涉及HP代谢、转录翻译、氧化还原和黏附等,并参与HP生存适应、定殖及致病过程。GroEL有望成为一种新的研究HP致病性及发展感染干预策略的重要靶点。
| [1] | SONNENBERG A, LASH RH, GENTA RM. A national study of Helicobactor pylori infection in gastric biopsy specimens. Gastroenterology, 2010, 139(6): 1894-1901.e2. DOI:10.1053/j.gastro.2010.08.018 |
| [2] | SIPPONEN P. Natural history of gastritis and its relationship to peptic ulcer disease. Digestion, 1992, 51(suppl 1): 70-75. |
| [3] | MOAYYEDI P, FORMAN D, BRAUNHOLTZ D, FELTBOWER R, CROCOMBE W, LIPTROTT M, AXON A. The proportion of upper gastrointestinal symptoms in the community associated with Helicobacter pylori, lifestyle factors, and nonsteroidal anti-inflammatory drugs. The American Journal of Gastroenterology, 2000, 95(6): 1448-1455. DOI:10.1111/j.1572-0241.2000.2126_1.x |
| [4] | SUGANO K. Screening of gastric cancer in Asia. Best Practice & Research Clinical Gastroenterology, 2015, 29(6): 895-905. |
| [5] | RONCARATI D, SCARLATO V. Roles and regulation of the heat shock proteins of the major human pathogen Helicobacter pylori[M]//Regulation of Heat Shock Protein Responses. Cham: Springer International Publishing. 2018: 411-427. |
| [6] | FERNÁNDEZ-DE-LARREA N, MICHEL A, ROMERO B, BUTT J, PAWLITA M, PÉREZ-GÓMEZ B, CASTAÑO-VINYALS G, MORENO V, MARTÍN V, AMIANO P, CASTILLA J, FERNÁNDEZ-TARDÓN G, DIERSSEN-SOTOS T, CLOFENT J, ALGUACIL J, HUERTA JM, JIMÉNEZ-MOLEÓN JJ, BARRICARTE A, MOLINUEVO A, FERNÁNDEZ-VILLA T, et al. Antibody reactivity against Helicobacter pylori proteins in a sample of the Spanish adult population in 2008‒2013. Helicobacter, 2017, 22(5). DOI:10.1111/hel.12401 |
| [7] | MICHEL A, PAWLITA M, BOEING H, GISSMANN L, WATERBOER T. Helicobacter pylori antibody patterns in Germany: a cross-sectional population study. Gut Pathogens, 2014, 6: 10. DOI:10.1186/1757-4749-6-10 |
| [8] | PAN KF, FORMICHELLA L, ZHANG L, ZHANG Y, MA JL, LI ZX, LIU C, WANG YM, GOETTNER G, ULM K, CLASSEN M, YOU WC, GERHARD M. Helicobacter pylori antibody responses and evolution of precancerous gastric lesions in a Chinese population. International Journal of Cancer, 2014, 134(9): 2118-2125. DOI:10.1002/ijc.28560 |
| [9] | GAO L, MICHEL A, WECK MN, ARNDT V, PAWLITA M, BRENNER H. Helicobacter pylori infection and gastric cancer risk: evaluation of 15 H. pylori proteins determined by novel multiplex serology. Cancer Research, 2009, 69(15): 6164-6170. DOI:10.1158/0008-5472.CAN-09-0596 |
| [10] | FORD PJ, GEMMELL E, HAMLET SM, HASAN A, WALKER PJ, WEST MJ, CULLINAN MP, SEYMOUR GJ. Cross-reactivity of GroEL antibodies with human heat shock protein 60 and quantification of pathogens in atherosclerosis. Oral Microbiology and Immunology, 2005, 20(5): 296-302. DOI:10.1111/j.1399-302X.2005.00230.x |
| [11] | EWALT KL, HENDRICK JP, HOURY WA, HARTL FU. In vivo observation of polypeptide flux through the bacterial chaperonin system. Cell, 1997, 90(3): 491-500. DOI:10.1016/S0092-8674(00)80509-7 |
| [12] | HOURY WA, FRISHMAN D, ECKERSKORN C, LOTTSPEICH F, HARTL FU. Identification of in vivo substrates of the chaperonin GroEL. Nature, 1999, 402(6758): 147-154. DOI:10.1038/45977 |
| [13] | PHADNIS SH, PARLOW MH, LEVY M, ILVER D, CAULKINS CM, CONNORS JB, DUNN BE. Surface localization of Helicobacter pylori urease and a heat shock protein homolog requires bacterial autolysis. Infection and Immunity, 1996, 64(3): 905-912. DOI:10.1128/iai.64.3.905-912.1996 |
| [14] | OLOFSSON A, VALLSTRÖM A, PETZOLD K, TEGTMEYER N, SCHLEUCHER J, CARLSSON S, HAAS R, BACKERT S, WAI SN, GRÖBNER G, ARNQVIST A. Biochemical and functional characterization of Helicobacter pylori vesicles. Molecular Microbiology, 2010, 77(6): 1539-1555. DOI:10.1111/j.1365-2958.2010.07307.x |
| [15] | RAUCH J, BARTON J, KWIATKOWSKI M, WUNDERLICH M, STEFFEN P, MODERZYNSKI K, PAPP S, HÖHN K, SCHWANKE H, WITT S, RICHARDT U, MEHLHOOP U, SCHLÜTER H, PIANKA V, FLEISCHER B, TAPPE D, OSTERLOH A. GroEL is an immunodominant surface-exposed antigen of Rickettsia typhi. PLoS One, 2021, 16(6): e0253084. DOI:10.1371/journal.pone.0253084 |
| [16] | BOTTERO D, ZURITA ME, GAILLARD ME, BARTEL E, VERCELLINI C, HOZBOR D. Membrane vesicles derived from Bordetella bronchiseptica: active constituent of a new vaccine against infections caused by this pathogen. Applied and Environmental Microbiology, 2018, 84(4): e01877-e01817. |
| [17] | MAHAWAR M, TRAN V, SHARP JS, MAIER RJ. Synergistic roles of Helicobacter pylori methionine sulfoxide reductase and GroEL in repairing oxidant-damaged catalase. Journal of Biological Chemistry, 2011, 286(21): 19159-19169. DOI:10.1074/jbc.M111.223677 |
| [18] | EVANS DJ Jr, EVANS DG, ENGSTRAND L, GRAHAM DY. Urease-associated heat shock protein of Helicobacter pylori. Infection and Immunity, 1992, 60(5): 2125-2127. DOI:10.1128/iai.60.5.2125-2127.1992 |
| [19] | ZHAO HL, WU YL, XU Z, MA R, DING YF, BAI XL, RONG QY, ZHANG Y, LI BQ, JI XF. Mechanistic insight into the interaction between Helicobacter pylori urease subunit α and its molecular chaperone Hsp60. Frontiers in Microbiology, 2019, 10: 153. DOI:10.3389/fmicb.2019.00153 |
 2023, Vol. 63
2023, Vol. 63




