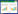中国科学院微生物研究所,中国微生物学会
文章信息
- 雷进田, 刘俊杰, 刘株秀, 梁爱珍, 胡晓婧, 于镇华, 金剑, 刘晓冰, 王光华. 2023
- LEI Jintian, LIU Junjie, LIU Zhuxiu, LIANG Aizhen, HU Xiaojing, YU Zhenhua, JIN Jian, LIU Xiaobing, WANG Guanghua.
- 保护性耕作增强了真菌群落生态网络稳定性
- Conservation tillage enhanced the stability of fungal community network
- 微生物学报, 63(7): 2835-2847
- Acta Microbiologica Sinica, 63(7): 2835-2847
-
文章历史
- 收稿日期:2022-11-01
- 网络出版日期:2023-01-05
2. 中国科学院大学, 北京 100049
2. University of Chinese Academy of Sciences, Beijing 100049, China
中国东北黑土区现有耕地面积3.59×107 ha,年粮食商品量占全国的1/4,调出量占全国的1/3,是我国重要的粮食生产基地[1-2]。然而长期的高强度利用和不合理耕作,如高密度的传统犁铧耕作(moldboard plow tillage, MP),导致了土壤侵蚀、土壤有机质含量下降以及生态功能退化等问题,严重影响了黑土区农业的可持续发展[3]。有研究证实保护性耕作的免耕措施(no-tillage, NT)可以有效地减缓土壤侵蚀、降低耕作成本以及减少农业灾害的发生[4-5]。然而,免耕容易造成土壤紧实度增加、种子出芽率降低、作物根系生长受限和病原微生物增加等一系列影响土壤健康和作物生长的不利因素[6-8]。与MP和NT相比,研究证实少耕(reduced tillage, RT)可以在一定程度上减少传统耕作和免耕的不利影响,具有维持土壤生态功能和作物产能协同提升的作用[9-10]。
土壤微生物通过调节土壤生物地球化学循环和植物养分吸收在农业生态系统中发挥着重要作用[11]。不同耕作方式改变了土壤性质、土壤微生物群落及相应的土壤生态功能[12]。传统耕作机械扰动下的土壤养分均一化导致土壤好氧微生物和r生存策略微生物群落迅速定殖,进而导致土壤微生物的生物量和多样性显著下降[13-14]。而保护性耕作在减少土壤扰动后致使土壤微生物由细菌型向真菌型转变[15-16]。此外,有研究表明长期免耕和秸秆覆盖会增加包括镰孢菌属(Fusarium)、腐霉属(Pythium)和丝核菌属(Rhizoctonia)等在内的植物潜在病原菌的数量[6, 17]。我国学者对免耕处理不同秸秆含量覆盖下土壤微生物群落变化的研究发现,与细菌群落相比,土壤真菌对秸秆覆盖响应更为敏感,秸秆覆盖显著增加了土壤镰孢菌属F. moniliforme和F. graminearum的相对丰度,进而导致免耕处理玉米根腐病的发生[18-19]。目前,关于保护性耕作措施下土壤微生物群落研究多偏重于微生物组成特征及分异规律,而功能微生物群落的变化特征,特别是聚焦真菌病原菌及病原菌与其关联微生物组的网络特征以及潜在生态功能研究相对较少[20]。
本研究采用高通量测序技术,对比研究传统犁耕(MP)和保护性耕作(NT和RT)下根际和非根际土壤真菌群落的组成关系,分析不同耕作措施下植物潜在病原菌的数量变化规律以及土壤病原真菌的生态网络特性。通过解析病原真菌生态网络中的关联微生物组,进而明确保护性耕作真菌群落的潜在生态功能,从微生物生态学角度为耕作措施的选用提供理论依据。
1 材料与方法 1.1 样地和样品采集本研究不同耕作措施试验布设在吉林省长春市中国科学院东北地理与农业生态研究所长期定位观测站(北纬44°59′,东经125°23′)。气候属于典型的温带大陆季风气候,年降水量为614 mm,年平均气温为6.4 ℃。土壤为典型黑土(typic hapludoll, USDA, 1993),土壤质地为黏壤土。试验设置少耕(RT)、免耕(NT)和犁耕(MP) 3种耕作措施,采用大豆-玉米的轮作种植模式,每个耕作措施包括4个随机重复,每块样地面积为25 m×7.8 m。
NT采用John Deere 7200免耕播种机播种,不采用任何土壤扰动措施;RT的土壤扰动仅比NT多了一次每年6月份的中耕起垄;MP的土壤扰动主要为作物收获后的犁耕(0–20 cm)、春季翻耕(7.5–10.0 cm)以及6月份的田垄修整。作物的收获季节为每年的10月份,大豆和玉米收获后进行秸秆还田。大豆统一施用的基肥量为40 kg/ha的氮、60 kg/ha的磷和80 kg/ha的钾。玉米施用的基肥量为100 kg/ha的氮、45.5 kg/ha的磷和78 kg/ha的钾,另外在玉米六叶期追加50 kg/ha的氮。采用混合施肥法,含有宏福缓释掺混肥(27% N, 12% P2O5, 13% K2O)、硝酸铵(NH4NO3)、过磷酸钙(CaP2H4O8)和硫酸钾(K2SO4)[12]。
本研究土壤采集于大豆-玉米轮作下的大豆茬口,非根际土壤随机选取3个土壤样本(0–20 cm),均匀混合后作为一个重复样本,每个处理随机取8个重复。此外,随机选取10株大豆,抖去肉眼可见的土壤颗粒后收集根际土壤样本[21]。本研究共采集土壤样品48份(3个处理×8重复×2根际/非根际)。冷藏土壤样本运回实验室后分为2部分,一部分保存于–80 ℃的冰箱,用于土壤DNA的提取及后续分子生物学分析;另一部分土壤风干后用于土壤理化性质的测定。
1.2 土壤理化性质的测定土壤pH测定采用pH计(Mettler Toledo, FE20 FiveEasyTM)在0.01 mol/L的CaCl2溶液中按照1:2.5 (土壤重量: 溶液体积)比例,振荡30 min后测得。土壤全碳(TC)和全氮(TN)利用VarioEL Ⅲ元素分析仪测定,取10 g土壤于2 mol/L的KCl溶液中振摇1 h后提取土壤铵盐(NH4+-N)和硝酸盐(NO3–-N)溶液。全磷(TP)和速效磷(AP)由H2SO4-HClO4消化法和NaHCO3萃取法分别提取。利用SKALAR San连续流动分析系统(Skalar)测定TP、AP、NH4+-N和NO3–-N的浓度。采用浓氢氟酸消化法提取土壤全钾(TK),速效钾(AK)使用乙酸铵浸提,通过电感耦合等离子体发射光谱法(ICPS-7500, Shimadze)测定TK和AK含量。
1.3 MiSeq测序以及生物信息学分析取0.5 g土样,使用土壤DNA提取试剂盒(Fast DNA® Spin Kit for Soil)按照试验操作要求进行土壤总DNA的提取,采用NanoDrop 2000 (Thermo Scientific)对提取的DNA进行浓度和质量分析。利用引物ITS1F (5′-CTTGGTCATTT AGAGGAAGTAA-3′)/ITS2R (5′-GCTGCGTTC TTCATCGATGC-3′)对真菌ITS区进行PCR扩增[22]。PCR产物采用Illumina MiSeq PE300平台进行真菌群落的高通量测序分析(Majorbio BioPharm Technology Co., Ltd., 上海美吉生物医药科技有限公司)。
使用QIIME (http://qiime.sourceforge.net/)平台对真菌原始数据进行序列拆分和质量控制。利用FLASH软件对质控后的序列进行拼接,并去除长度小于150 bp和平均质量小于20的序列信息,利用Uchime algorithm软件去除嵌合体序列[23-24]。通过RDP (ribosomal database project)对质控后的数据进行分类注释,去除非真菌序列后通过UPARSE基于97%的相似度进行真菌分类单元(operational taxonomic units, OTUs)[25]划分。本研究共获得1 436 544个高质量真菌序列,划分为1 831个真菌OTUs。原始序列以登录号PRJNA 644753保存在NCBI序列读取档案(SRA)中。
1.4 数据处理及分析本研究基于获得最少序列数将不同耕作措施的OTUs序列随机抽平到29 923条,以减少各处理序列数据不均造成的分析误差。利用FUNGuild工具(http://www.funguild.org/)对真菌OTUs进行营养级功能注释[26],筛选FUNGuild工具中注释的植物病原真菌绘制病原真菌相对丰度柱状图。为满足正态性和同质性方差检验,使用SPSS (Version 22.0)软件显著性水平P < 0.05的最小显著性差异(least-significant difference, LSD)分析土壤病原真菌的差异显著性。不同耕作措施下真菌群落的α多样性采用Shannon和PD (faith’s phylogenetic distance)指数进行表征,beta多样性及其驱动因素由真菌群落的主坐标分析(principal coordinate analysis, PCoA)和冗余分析(redundancy analysis, RDA)进行解析。Beta多样性的显著性使用多元方差分析(adonis)验证;PCoA利用R软件的“vegan”和“ggplot2”包进行分析和可视化处理,RDA分析前利用R软件的“car”包进行方差膨胀系数(variance inflation factor, VIF)检验,去除VIF > 10的环境因子[27]。利用R软件的“WGCNA”包对相对丰度高于0.1%的OTUs进行网络构建[28],使用“igraph”包对构建的真菌网络进行网络属性分析。利用Cytoscape (http://www.cytoscape.org/)进行土壤真菌网络可视化[29]。本研究不同处理共构建6个真菌病原菌网络,结合FUNGuild获得的真菌功能注释信息从真菌网络中抽取病原真菌子网络,并比较不同处理间的子网络拓扑性质。
2 结果与分析 2.1 不同耕作措施的土壤理化性质不同耕作下根际和非根际土壤理化性质如表 1[30]所示,与保护性耕作NT和RT相比,在根际和非根际土壤中MP均显著增加了土壤pH值。与NT对比,在根际和非根际土壤中MP和RT显著增加了土壤TC、TN和AK含量,而MP处理增加了硝态氮(nitrate nitrogen, NO3–-N)的含量。根际土壤中,MP处理下碳氮比(carbonnitrogen ratio, C/N)、全磷(total phosphorus, TP)和速效磷(available phosphorus, AP)的含量显著高于NT和RT,而NT显著增加了土壤全钾(Total potassium, TK)含量;在非根际土壤中,相比于NT和MP,RT处理显著增加了TP和AP含量,NT处理中TK的含量显著高于MP。
| Soil properties | NTB | RTB | MPB | NTR | RTR | MPR |
| pH | 5.47±0.01b | 5.46±0.03b | 5.75±0.04a | 5.41±0.01b | 5.41±0.01b | 5.70±0.03a |
| Total carbon (g/kg) | 14.1±0.39b | 18.3±0.41a | 18.7±0.45a | 19.2±0.20c | 25.5±1.09b | 26.5±0.68a |
| Total nitrogen (g/kg) | 1.66±0.09b | 2.03±0.11a | 2.19±0.23a | 2.15±0.05b | 2.72±0.06a | 2.68±0.07a |
| Carbon/Nitrogen | 8.54±0.43a | 9.06±0.65a | 8.58±0.78a | 8.95±0.18c | 9.38±0.46b | 9.90±0.40a |
| Total phosphorus (g/kg) | 0.45±0.00b | 0.47±0.01a | 0.45±0.02b | 0.47±0.02c | 0.51±0.01b | 0.54±0.01a |
| Total potassium (g/kg) | 23.4±0.21a | 23.3±0.21ab | 23.2±0.22b | 23.8±0.29a | 23.3±0.24b | 23.1±0.23b |
| Available phosphorus (mg/kg) | 15.2±1.10b | 19.6±0.51a | 15.4±0.23b | 18.2±1.25c | 23.5±0.56b | 27.2±1.45a |
| Available potassium (mg/kg) | 107±2.6c | 137±1.7a | 133±4.0b | 188±1.3b | 214±1.7a | 213±1.2a |
| NH4+-N (mg/kg) | 1.71±0.18a | 1.75±0.19a | 1.78±0.19a | 3.45±0.36a | 3.47±0.40a | 3.17±0.39a |
| NO3‒-N (mg/kg) | 0.20±0.06c | 0.27±0.05b | 0.37±0.07a | 0.83±0.12b | 0.88±0.16b | 1.33±0.22a |
| The values are means of eight replicates, with different letters indicating significant differences (P < 0.05) among tillage practices for bulk or rhizosphere soil. NT: No-tillage; RT: Reduced tillage; MP: Moldboard plow tillage; B: Bulk soils; R: Rhizosphere soils. | ||||||
2.2 不同耕作措施的真菌群落α和β多样性
非根际土壤中不同耕作措施下Shannon指数和PD值不存在显著差异;在根际土壤中,RT处理的Shannon指数显著高于MP和NT,RT的PD值显著高于NT处理(图 1A、1B)。基于Bray-Curtis距离的PCoA分析结果显示(图 1C),真菌群落结构显著被分为非根际和根际土壤2个集群,而在根际和非根际土壤内,不同耕作方式真菌群落结构间均存在显著差异(Adonis, P < 0.001)。此外,不同耕作措施真菌群落的差异度在根际土壤中要显著高于非根际土壤。RDA结果显示,土壤pH和土壤养分(包括TC、TP、TK和铵态氮(ammonium nitrogen,NH4+-N)是不同耕作措施真菌群落发生分异的主要驱动因子(图 1D) (R2=0.50, P < 0.01)。
|
| 图 1 不同耕作措施下根际和非根际土壤真菌的Shannon指数(A)、系统发育多样性(PD) (B)、主坐标分析(PCoA) (C)和冗余分析(RDA) (D) Figure 1 Shannon's index (A), phylogenetic diversity (PD) (B), principal coordinate analysis (PCoA) (C) and redundancy analysis (RDA) (D) of rhizosphere and bulk soils under different tillage measures. |
2.3 不同耕作措施对土壤真菌群落组成的影响
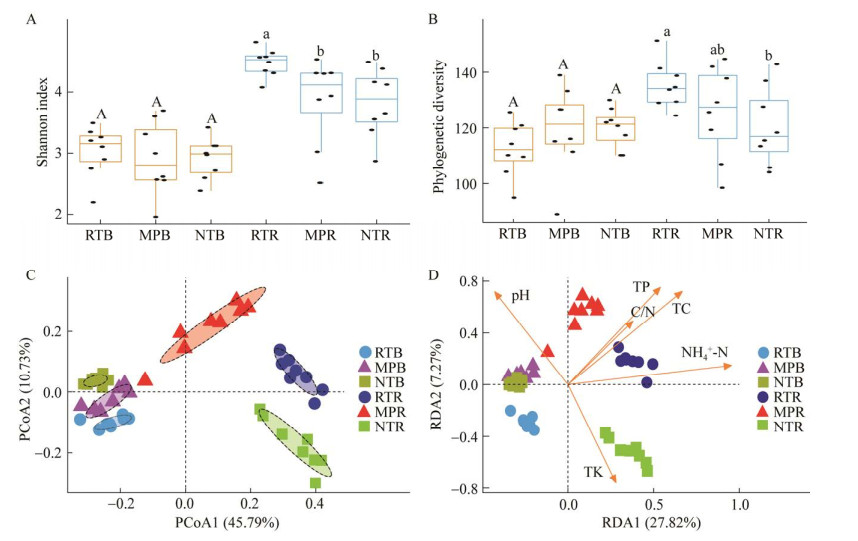
本研究共检测出7个真菌门类,在非根际土壤中,担子菌门(Basidiomycota)为优势真菌门,相对丰度为63.1%–70.1%,其次是子囊菌门(Ascomycota),相对丰度为23.9%–25.5%;而根际土壤内,子囊菌门为优势真菌门,相对丰度为55.2%–58.7%,其次为担子菌门,相对丰度为23.8%–31.8%。罗兹菌门(Rozellomycota)和球囊菌门(Glomeromycota)为痕量真菌类群,其相对丰度占比0.5%左右。非根际土壤中,与NT和RT相比,MP显著增加了担子菌门的相对丰度;根际土壤中,与MP和NT对比,RT显著降低了担子菌门丰度,显著增加了子囊菌门的相对丰度(图 2A)。
|
| 图 2 不同耕作措施下真菌门相对丰度堆叠柱状图(A)和病原真菌相对丰度柱状图(B) Figure 2 Percentage histogram of phylum (A) and pathogenic fungi (B) under different tillage measures. |
非根际土壤中,尽管不同耕作措施下的病原菌相对丰度不存在显著差异,但是RT处理下病原菌相对丰度依旧略低于NT和MP;在根际土壤中,MP处理显著(P < 0.01)增加了病原真菌的相对丰度(3.64%),分别高于RT和NT处理的2.52%和2.81% (图 2B)。
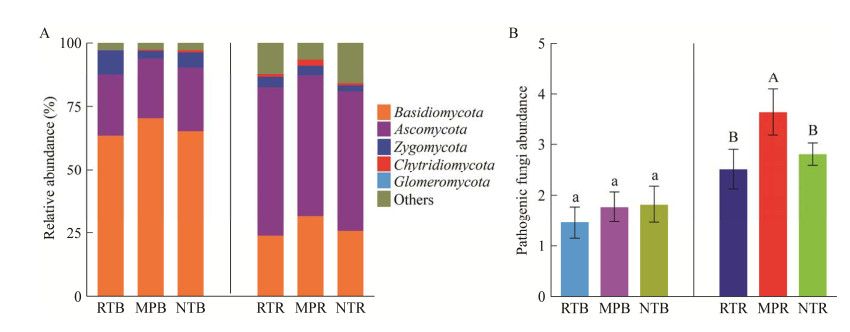
基于真菌群落OTUs水平的Venn图分析结果显示,在根际和非根际土壤中,RT独有的OTUs数均显著高于MP和NT (图 3A),不同的耕作措施之间也存在显著的差异,保护性耕作RT和NT处理的OTUs显著高于MP。三元相图(图 3B)结果显示,耕作方式对于真菌OTUs的数量具有显著影响,根际土壤中RT处理下的OTUs数量显著高于MP和NT,非根际土壤中,RT和MP处理的OTUs数量显著高于NT。对比耕作方式真菌OTUs变化特征发现,10个相对丰度较高的OTUs中包含OTU1075 (Fusarium brachygibbosum)和OTU1803 (Alternaria) 2种大豆潜在病原真菌(图 3C),并且NT和RT显著降低了根际土壤中OTU1075和OTU1803的相对丰度。
|
| 图 3 不同耕作下根际和非根际土壤真菌的Venn图(A)、三元相图(B)和优势真菌相对丰度(C) Figure 3 Venn diagram (A), ternary phase diagrams (B), and dominant fungi relative abundance plots (C) of bulk and rhizosphere soils under different tillage measures. |
2.4 不同耕作措施下土壤病原菌生态网络
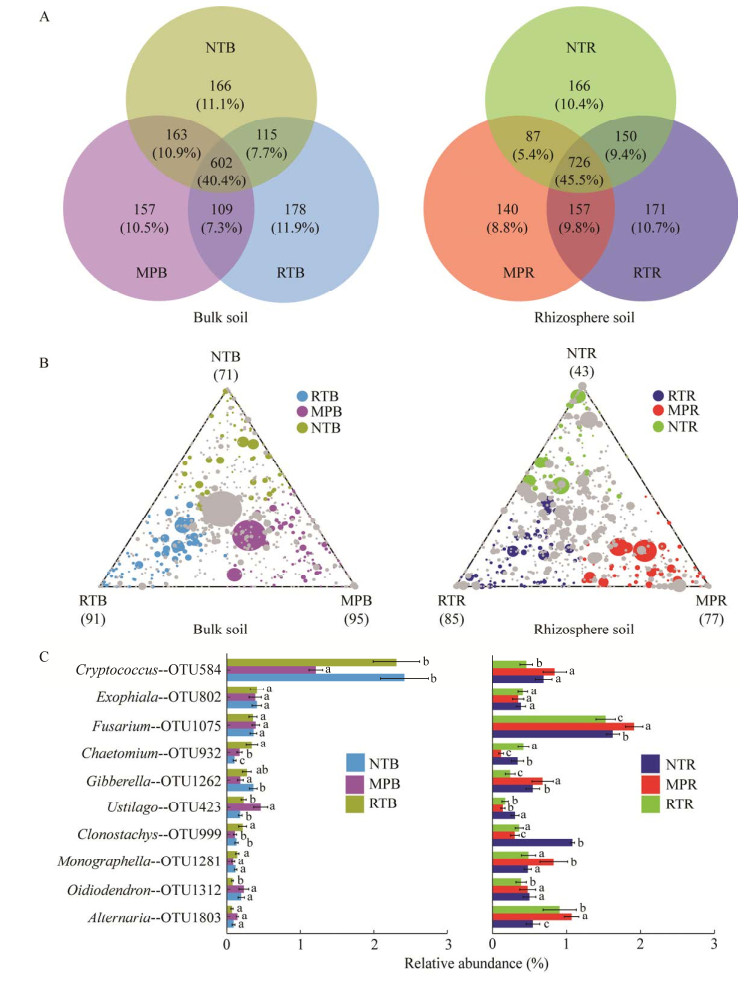
不同耕作措施下土壤潜在植物病原真菌与其关联真菌群落的生态网络结果显示(图 4),在根际和非根际土壤中,MP处理下土壤病原真菌的节点数(nodes)、连线数(edges)和以及与关联真菌之间的平均连接度(average degree)均显著高于RT和NT,但是显著降低了网络模块度(modularity);而RT处理植物病原真菌的nodes、edges和average degree值要低于NT和MP处理,尤其在根际土壤中。此外,病原真菌与关联真菌间的交互作用中85%为正交互作用关系,其中在根际和非根际土壤中,MP处理病原真菌与关联真菌间的正交互作用大于94%,显著高于NT和RT。
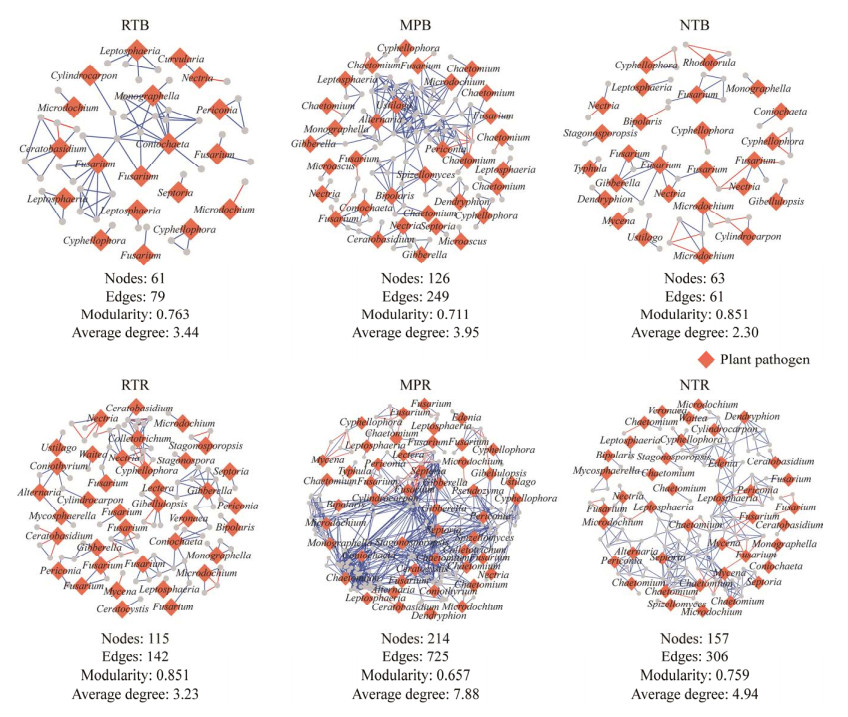
|
| 图 4 不同耕作措施下根际和非根际土壤病原真菌的生态网络图 Figure 4 The ecological network plots of bulk and rhizosphere pathogenic fungi under different tillage measures. Blue lines: Positive correlation; Red lines: Negative correlation. |
3 讨论
以往研究证实,与传统耕作措施相比,保护性耕作NT显著增加土壤养分含量,而保护性耕作下土壤养分含量的增加主要是由秸秆还田直接导致而非耕作措施本身引起的[31-32]。本研究发现NT处理在根际和非根际土壤中均显著降低了土壤TC和TN含量(表 1),这与之前研究结果相悖[33],主要原因在于本研究中不同耕作措施均还田了相同的秸秆量。有研究表明秸秆覆盖下的NT处理可以增加腐生真菌数量[3-4, 20],而腐生真菌的菌丝网络可以充分利用秸秆碳和土壤中的氮元素[19];其次本研究的非根际土壤是采集0–20 cm的土壤均匀混合样本,因此NT处理下的秸秆表面覆盖方式并未导致耕层土壤养分含量的增加。Zhang等[34]在对比秸秆还田下保护性耕作不同深度土壤养分含量变化的研究中发现,与MP对比,在0–5 cm土层NT处理SOC含量显著增加,而在5–20 cm土层NT与MP处理间差异不显著。本研究发现与NT和RT相比,MP处理显著增加了土壤pH值(表 1),而已有研究结果证实保护性耕作对土壤pH的影响并无一致性的变化规律,土壤pH主要受到土壤类型、气候条件和管理因素等多因素影响[35-36],且其变化并不是直接由耕作措施引起的[30]。
土壤微生物多样性与土壤肥力、作物产量及农业生态系统功能稳定性等因素密切关联[37-38],高的土壤微生物多样性对群落功能、作物生长等具有促进作用,同时对于病原菌的入侵和定殖具有一定的抵抗作用[39-41]。本研究发现,在根际土壤中保护性耕作NT和RT特有的真菌数量(OTUs)显著高于传统耕作MP处理(图 3A),且RT处理的Shannon指数显著高于NT和MP (图 1A),表明保护性耕作有利于增加土壤微生物多样性和物种丰富度,增强微生物间的相互作用和微生物群落功能、促进作物生长[38],从而抑制土壤中植物病原菌的入侵和定殖。有研究证实,秸秆还田下的NT和RT处理会增加土壤中微生物以及真菌丰富度[4, 19]。本研究发现,MP处理在根际土壤中显著增加了如OTU1803 (Alternaria)等大豆病原菌的相对丰度。其主要原因是在相同秸秆还田量下的3种耕作措施中,MP处理将农田中的土壤均匀翻耕的同时将有机质均匀地分散在整个耕层中,促使土壤形成比表面积更大且结构更加合理的团聚体,增强了病原菌的定植和养分利用率[22, 42]。然而相比于MP,RT处理显著降低了根际土壤中OTU1075 (Fusarium brachygibbosum)等大豆病原菌的相对丰度(图 3C),间接佐证了RT处理多样性指数高且具有较低丰度病原菌的结论。此外有研究证实,保护性耕作NT和RT通过增加土壤水分的可用性、维持土壤温度、提高土壤养分含量和促进真菌形成菌丝网络等作用,进而增加土壤真菌的活性[3-4, 18]。如Wang等[19]的研究证实,保护性耕作NT和RT处理显著增加了毛壳菌属(Chaetomium)和葡萄穗霉属(Stachybotrys)等真菌数量,而毛壳菌属对植物病原体黄色镰孢菌(Fusarium culmorum)和致病疫霉(Phytophthora infestans)等具有很强的抑制作用[43];葡萄穗霉属(Stachybotrys elegans)通过寄生抑制立枯丝核菌(Rhizoctonia solani)病原菌的增长[44]。
本研究发现不同耕作措施下土壤真菌群落存在显著差异(Adonis, P=0.001),土壤pH和土壤养分含量是导致不同耕作措施真菌群落发生分异的最主要驱动因子(图 1D)。有研究证实这是由于不同耕作措施可以诱导土壤理化性质的变化,塑造不同的土壤微生物生存环境,从而导致土壤微生物群落结构的差异[45-46]。研究发现土壤真菌群落受到根际效应的影响要强于耕作措施(图 1C),这主要是根际环境中根际分泌物和沉积物的富集导致的[21],并且在根际土壤中耕作方式引起的真菌群落结构的差异也远大于非根际土壤,这主要是由于根际土壤中拥有更高的微生物丰富度和多样性[38],且根际土壤微生物对环境变化更敏感[47]。以往研究证实根际真菌在抵抗病原菌侵染过程中发挥重要作用[48],然而本研究发现,相同耕作措施下,根际土壤病原菌的相对丰度显著高于非根际土壤(图 2B)。其主要原因是长期连续的耕作下,根际真菌对病原菌的抵抗作用会丧失[49],其次,根际环境中根际分泌物的种类和数量要高于非根际土壤,如氨基酸、有机酸等分泌物可被病原真菌直接利用[22, 47, 50],为根际土壤中病原真菌的生长提供了能量和养分。
Six等[51]和Le Guillou等[52]的研究证实,MP的均匀翻耕促进了真菌对土壤养分的利用,加强了真菌间的合作。Hu等[22]对不同耕作措施下土壤细菌、真菌和古菌群落的生态网络分析发现,MP处理导致了病原真菌与其他真菌群落之间更多的正相互作用。这进一步证实了我们的研究结果,MP处理的网络节点数和网络复杂度均显著高于NT和RT (图 4)。有研究将模块定义为生态位[45],并且用网络模块性来表征网络结构的稳定性,模块性越大其代表的网络稳定性越高[12, 53-54]。本研究中,相比于MP,RT处理具有更高的模块性且病原真菌与其他真菌群落间具有更低的连接性(图 4),说明RT处理在提升真菌群落网络稳定性的同时降低了生态网络中潜在植物病原真菌与其他关联真菌之间的合作,这有利于减缓土壤中潜在植物病原菌的生态影响,降低潜在植物病原菌在微生物网络中的转移效率。
4 结论不同耕作措施显著改变了土壤理化性状。与NT处理对比,MP和RT显著增加了土壤养分含量,这主要是由于在相同秸秆量还田下土壤表面秸秆覆盖还田的NT对土壤养分贡献显著低于扰动后秸秆被均匀翻混到耕层的MP和RT处理。根际效应对真菌群落的影响显著高于耕作措施,不同耕作措施下真菌群落多样性指数、群落结构组成及病原菌丰度变化在根际土壤中均显著强于非根际土壤。MP处理均匀翻耕土壤促使土壤形成比表面积更大且结构更加合理的团聚体,显著增加了链格孢霉菌和镰孢菌的相对丰度。MP处理增强了病原真菌与关联真菌网络的正相互作用,而RT处理提升了真菌群落网络稳定性,同时降低了生态网络中潜在植物病原真菌的传播,因而是具有提升土壤质量和作物产能的耕作措施。
| [1] |
李保国, 刘忠, 黄峰, 杨晓光, 刘志娟, 万炜, 汪景宽, 徐英德, 李子忠, 任图生. 巩固黑土地粮仓保障国家粮食安全[J]. 中国科学院院刊, 2021, 36(10): 1184-1193.
LI BG, LIU Z, HUANG F, YANG XG, LIU ZJ, WAN W, WANG JK, XU YD, LI ZZ, REN TS. Ensuring national food security by strengthening high-productivity black soil granary in Northeast China[J]. Bulletin of Chinese Academy of Sciences, 2021, 36(10): 1184-1193 (in Chinese). |
| [2] |
陈玉洁, 张平宇, 刘世薇, 谭俊涛. 东北西部粮食生产时空格局变化及优化布局研究[J]. 地理科学, 2016, 36(9): 1397-1407.
DOI:10.13249/j.cnki.sgs.2016.09.013 CHEN YJ, ZHANG PY, LIU SW, TAN JT. The spatio-temporal pattern change and optimum layout of grain production in the west of Northeast China[J]. Scientia Geographica Sinica, 2016, 36(9): 1397-1407 (in Chinese). |
| [3] | ZHANG B, HE HB, DING XL, ZHANG XD, ZHANG XP, YANG XM, FILLEY TR. Soil microbial community dynamics over a maize (Zea mays L.) growing season under conventional- and no-tillage practices in a rainfed agroecosystem[J]. Soil & Tillage Research, 2012, 124: 153-160. |
| [4] | BUSARI MA, KUKAL SS, KAUR A, BHATT R, DULAZI AA. Conservation tillage impacts on soil, crop and the environment[J]. International Soil and Water Conservation Research, 2015, 3(2): 119-129 DOI:10.1016/j.iswcr.2015.05.002. |
| [5] | ZHAO X, LIU SL, PU C, ZHANG XQ, XUE JF, REN YX, ZHAO XL, CHEN F, LAL R, ZHANG HL. Crop yields under no-till farming in China: a meta-analysis[J]. European Journal of Agronomy, 2017, 84: 67-75 DOI:10.1016/j.eja.2016.11.009. |
| [6] | GOVAERTS B, MEZZALAMA M, SAYRE KD, CROSSA J, NICOL JM, DECKERS J. Long-term consequences of tillage, residue management, and crop rotation on maize/wheat root rot and nematode populations in subtropical highlands[J]. Applied Soil Ecology, 2006, 32(3): 305-315 DOI:10.1016/j.apsoil.2005.07.010. |
| [7] | BAILEY A, DEASY C, QUINTON J, SILGRAM M, JACKSON B, STEVENS C. Determining the cost of in-field mitigation options to reduce sediment and phosphorus loss[J]. Land Use Policy, 2013, 30(1): 234-242 DOI:10.1016/j.landusepol.2012.03.027. |
| [8] | DANG YP, SEYMOUR NP, WALKER SR, BELL MJ, FREEBAIRN DM. Strategic tillage in no-till farming systems in Australia's northern grains-growing regions: i. drivers and implementation[J]. Soil & Tillage Research, 2015, 152: 104-114. |
| [9] | ALVAREZ R. A review of nitrogen fertilizer and conservation tillage effects on soil organic carbon storage[J]. Soil Use and Management, 2005, 21(1): 38-52 DOI:10.1079/SUM2005291. |
| [10] | HOBBS PR, SAYRE K, GUPTA R. The role of conservation agriculture in sustainable agriculture[J]. Philosophical Transactions of the Royal Society B: Biological Sciences, 2008, 363(1491): 543-555 DOI:10.1098/rstb.2007.2169. |
| [11] | FALKOWSKI PG, FENCHEL T, DELONG EF. The microbial engines that drive earth's biogeochemical cycles[J]. Science, 2008, 320(5879): 1034-1039 DOI:10.1126/science.1153213. |
| [12] | LIU ZX, GU HD, LIANG AZ, LI LJ, YAO Q, XU YX, LIU JJ, JIN J, LIU XB, WANG GH. Conservation tillage regulates the assembly, network structure and ecological function of the soil bacterial community in black soils[J]. Plant and Soil, 2021, 472(1/2): 207-223. |
| [13] | WANG ZT, LIU L, CHEN Q, WEN XX, LIAO YC. Conservation tillage increases soil bacterial diversity in the dryland of northern China[J]. Agronomy for Sustainable Development, 2016, 36(2): 28 DOI:10.1007/s13593-016-0366-x. |
| [14] | ZUBER SM, VILLAMIL MB. Meta-analysis approach to assess effect of tillage on microbial biomass and enzyme activities[J]. Soil Biology & Biochemistry, 2016, 97: 176-187. |
| [15] | ACOSTA-MARTÍNEZ V, MIKHA MM, VIGIL MF. Microbial communities and enzyme activities in soils under alternative crop rotations compared to wheat-fallow for the Central Great Plains[J]. Applied Soil Ecology, 2007, 37(1/2): 41-52. |
| [16] | MINOSHIMA H, JACKSON LE, CAVAGNARO TR, SÁNCHEZ-MORENO S, FERRIS H, TEMPLE SR, GOYAL S, MITCHELL JP. Soil food webs and carbon dynamics in response to conservation tillage in California[J]. Soil Science Society of America Journal, 2007, 71(3): 952-963 DOI:10.2136/sssaj2006.0174. |
| [17] | ROGET DK, NEATE SM, ROVIRA AD. Effect of sowing point design and tillage practice on the incidence of rhizoctonia root rot, take-all and cereal cyst nematode in wheat and barley[J]. Australian Journal of Experimental Agriculture, 1996, 36(6): 683-693 DOI:10.1071/EA9960683. |
| [18] | WANG HH, LI X, LI X, WANG J, LI XY, GUO QC, YU ZX, YANG TT, ZHANG HW. Long-term no-tillage and different residue amounts alter soil microbial community composition and increase the risk of maize root rot in Northeast China[J]. Soil & Tillage Research, 2020, 196: 104452. |
| [19] | WANG HH, GUO QC, LI X, LI X, YU ZX, LI XY, YANG TT, SU ZC, ZHANG HW, ZHANG CG. Effects of long-term no-tillage with different straw mulching frequencies on soil microbial community and the abundances of two soil-borne pathogens[J]. Applied Soil Ecology, 2020, 148: 103488 DOI:10.1016/j.apsoil.2019.103488. |
| [20] | ZHANG CF, LIN ZL, QUE YX, FALLAH N, TAYYAB M, LI SY, LUO J, ZHANG ZC, ABUBAKAR AY, ZHANG H. Straw retention efficiently improves fungal communities and functions in the fallow ecosystem[J]. BMC Microbiology, 2021, 21(1): 52 DOI:10.1186/s12866-021-02115-3. |
| [21] | PHILIPPOT L, RAAIJMAKERS JM, LEMANCEAU P, van der PUTTEN WH. Going back to the roots: the microbial ecology of the rhizosphere[J]. Nature Reviews Microbiology, 2013, 11(11): 789-799 DOI:10.1038/nrmicro3109. |
| [22] | HU XJ, LIU JJ, LIANG AZ, LI LJ, YAO Q, YU ZH, LI YS, JIN J, LIU XB, WANG GH. Conventional and conservation tillage practices affect soil microbial co-occurrence patterns and are associated with crop yields[J]. Agriculture Ecosystems & Environment, 2021, 319: 107534. |
| [23] | CAPORASO JG, LAUBER CL, WALTERS WA, BERG-LYONS D, HUNTLEY J, FIERER N, OWENS SM, BETLEY J, FRASER L, BAUER M, GORMLEY N, GILBERT JA, SMITH G, KNIGHT R. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms[J]. The ISME Journal, 2012, 6(8): 1621-1624 DOI:10.1038/ismej.2012.8. |
| [24] | TEDERSOO L, TOOMING-KLUNDERUD A, ANSLAN S. PacBio metabarcoding of fungi and other eukaryotes: errors, biases and perspectives[J]. The New Phytologist, 2018, 217(3): 1370-1385 DOI:10.1111/nph.14776. |
| [25] | EDGAR RC. UPARSE: highly accurate OTU sequences from microbial amplicon reads[J]. Frontiers in Plant Science, 2013, 10(10): 996-998. |
| [26] | NGUYEN NH, SONG ZW, BATES ST, BRANCO S, TEDERSOO L, MENKE J, SCHILLING JS, KENNEDY PG. FUNGuild: an open annotation tool for parsing fungal community datasets by ecological guild[J]. Fungal Ecology, 2016, 20: 241-248 DOI:10.1016/j.funeco.2015.06.006. |
| [27] | LI HY, QIU YZ, YAO T, HAN DR, GAO YM, ZHANG JG, MA YC, ZHANG HR, YANG XL. Nutrients available in the soil regulate the changes of soil microbial community alongside degradation of alpine meadows in the northeast of the Qinghai-Tibet Plateau[J]. The Science of the Total Environment, 2021, 792: 148363 DOI:10.1016/j.scitotenv.2021.148363. |
| [28] | FRIEDMAN J, ALM EJ. Inferring correlation networks from genomic survey data[J]. PLoS Computational Biology, 2012, 8(9): e1002687 DOI:10.1371/journal.pcbi.1002687. |
| [29] | SHANNON P, MARKIEL A, OZIER O, BALIGA NS, WANG JT, RAMAGE D, AMIN N, SCHWIKOWSKI B, IDEKER T. Cytoscape: a software environment for integrated models of biomolecular interaction networks[J]. Frontiers in Immunology, 2003, 13(11): 2498-2504. |
| [30] | HU XJ, LIANG AZ, YAO Q, LIU ZX, YU ZH, WANG GH, LIU JJ. Ridge tillage improves soil properties, sustains diazotrophic communities, and enhances extensively cooperative interactions among diazotrophs in a clay loam soil[J]. Frontiers in Microbiology, 2020, 11: 1333 DOI:10.3389/fmicb.2020.01333. |
| [31] | SUN BJ, JIA SX, ZHANG SX, MCLAUGHLIN NB, LIANG AZ, CHEN XW, LIU SY, ZHANG XP. No tillage combined with crop rotation improves soil microbial community composition and metabolic activity[J]. Environmental Science and Pollution Research International, 2016, 23(7): 6472-6482 DOI:10.1007/s11356-015-5812-9. |
| [32] | MELERO S, PANETTIERI M, MADEJON E, MACPHERSON HG, MORENO F, MURILLO JM. Implementation of chiselling and mouldboard ploughing in soil after 8 years of no-till management in SW, Spain: effect on soil quality[J]. Soil & Tillage Research, 2011, 112(2): 107-113. |
| [33] | CHEN HH, DAI ZM, VEACH AM, ZHENG JQ, XU JM, SCHADT CW. Global meta-analyses show that conservation tillage practices promote soil fungal and bacterial biomass[J]. Agriculture Ecosystems & Environment, 2020, 293: 106841. |
| [34] | ZHANG Y, LI XJ, GREGORICH EG, MCLAUGHLIN NB, ZHANG XP, GUO YF, GAO Y, LIANG AZ. Evaluating storage and pool size of soil organic carbon in degraded soils: tillage effects when crop residue is returned[J]. Soil and Tillage Research, 2019, 192: 215-221 DOI:10.1016/j.still.2019.05.013. |
| [35] | COOKSON WR, MURPHY DV, ROPER MM. Characterizing the relationships between soil organic matter components and microbial function and composition along a tillage disturbance gradient[J]. Soil Biology & Biochemistry, 2008, 40(3): 763-777. |
| [36] | RAHMAN MH, OKUBO A, SUGIYAMA S, MAYLAND HF. Physical, chemical and microbiological properties of an andisol as related to land use and tillage practice[J]. Soil & Tillage Research, 2008, 101(1/2): 10-19. |
| [37] | WAGG C, BENDER SF, WIDMER F, van der HEIJDEN MG. Soil biodiversity and soil community composition determine ecosystem multifunctionality[J]. Proceedings of the National Academy of Sciences of the United States of America, 2014, 111(14): 5266-5270 DOI:10.1073/pnas.1320054111. |
| [38] | SALEEM M, HU J, JOUSSET A. More than the sum of its parts: microbiome biodiversity as a driver of plant growth and soil health[J]. Annual Review of Ecology, Evolution, and Systematics, 2019, 50(1): 145-168 DOI:10.1146/annurev-ecolsys-110617-062605. |
| [39] | SCHMIDT R, MITCHELL J, SCOW K. Cover cropping and no-till increase diversity and symbiotroph: saprotroph ratios of soil fungal communities[J]. Soil Biology & Biochemistry, 2019, 129: 99-109. |
| [40] | van BRUGGEN AH, FINCKH MR. Plant diseases and management approaches in organic farming systems[J]. Annual Review of Phytopathology, 2016, 54: 25-54 DOI:10.1146/annurev-phyto-080615-100123. |
| [41] | LARKIN RP. Soil health paradigms and implications for disease management[J]. Annual Review of Phytopathology, 2015, 53: 199-221 DOI:10.1146/annurev-phyto-080614-120357. |
| [42] | XIA Q, LIU XL, GAO ZQ, WANG JM, YANG ZP. Responses of rhizosphere soil bacteria to 2-year tillage rotation treatments during fallow period in semiarid southeastern Loess Plateau[J]. PeerJ, 2020, 8: e8853 DOI:10.7717/peerj.8853. |
| [43] | LINKIES A, JACOB S, ZINK P, MASCHEMER M, MAIER W, KOCH E. Characterization of cultural traits and fungicidal activity of strains belonging to the fungal genus Chaetomium[J]. Journal of Applied Microbiology, 2021, 131(1): 375-391 DOI:10.1111/jam.14946. |
| [44] | BENYAGOUB M, JABAJI-HARE SH, BANVILLE G, CHAREST PM. Stachybotrys elegans: a destructive mycoparasite of Rhizoctonia solani[J]. Mycological Research, 1994, 98(5): 493-505 DOI:10.1016/S0953-7562(09)80467-X. |
| [45] | WANG Q, LIANG AZ, CHEN XW, ZHANG SX, ZHANG Y, MCLAUGHLIN NB, GAO Y, JIA SX. The impact of cropping system, tillage and season on shaping soil fungal community in a long-term field trial[J]. European Journal of Soil Biology, 2021, 102: 103253 DOI:10.1016/j.ejsobi.2020.103253. |
| [46] | NAVARRO-NOYA YE, GOMEZ-ACATA S, MONTOYA-CIRIACO N, ROJAS-VALDEZ A, SUAREZ-ARRIAGA MC, VALENZUELA-ENCINAS C, JIMENEZ-BUENO N, VERHULST N, GOVAERTS B, DENDOOVEN L. Relative impacts of tillage, residue management and crop-rotation on soil bacterial communities in a semi-arid agroecosystem[J]. Soil Biology & Biochemistry, 2013, 65: 86-95. |
| [47] | ZHANG BG, ZHANG J, LIU Y, SHI P, WEI GH. Co-occurrence patterns of soybean rhizosphere microbiome at a continental scale[J]. Soil Biology & Biochemistry, 2018, 118: 178-186. |
| [48] | EHRMANN J, RITZ K. Plant: soil interactions in temperate multi-cropping production systems[J]. Plant and Soil, 2013, 376(1/2): 1-29. |
| [49] | MAPELLI F, MARASCO R, FUSI M, SCAGLIA B, TSIAMIS G, ROLLI E, FODELIANAKIS S, BOURTZIS K, VENTURA S, TAMBONE F, ADANI F, BORIN S, DAFFONCHIO D. The stage of soil development modulates rhizosphere effect along a High Arctic Desert chronosequence[J]. The ISME Journal, 2018, 12(5): 1188-1198 DOI:10.1038/s41396-017-0026-4. |
| [50] | JIANG YJ, LI SZ, LI RP, ZHANG J, LIU YH, LV LF, ZHU H, WU WL, LI WL. Plant cultivars imprint the rhizosphere bacterial community composition and association networks[J]. Soil Biology & Biochemistry, 2017, 109: 145-155. |
| [51] | SIX J, PAUSTIAN K, ELLIOTT ET, COMBRINK C. Soil structure and organic matter Ⅰ. distribution of aggregate-size classes and aggregate-associated carbon[J]. Soil Science Society of America Journal, 2000, 64(2): 681-689. |
| [52] | LE GUILLOU C, ANGERS DA, LETERME P, MENASSERI-AUBRY S. Changes during winter in water-stable aggregation due to crop residue quality[J]. Soil Use and Management, 2012, 28(4): 590-595. |
| [53] | KITANO H. Biological robustness[J]. Nature Reviews Genetics, 2004, 5(11): 826-837. |
| [54] | DENG YL, RUAN YJ, MA B, TIMMONS MB, LU HF, XU XY, ZHAO HP, YIN XW. Multi-omics analysis reveals niche and fitness differences in typical denitrification microbial aggregations[J]. Environment International, 2019, 132: 105085. |
 2023, Vol. 63
2023, Vol. 63