中国科学院微生物研究所,中国微生物学会
文章信息
- 赵敏, 欧阳紫柔, 赵建宏. 2023
- ZHAO Min, OUYANG Zirou, ZHAO Jianhong.
- 艰难拟梭菌毒素致病机制及毒素基因调控的研究进展
- Advances in pathogenesis of Clostridioides difficile and regulation of toxin gene expression
- 微生物学报, 63(8): 2935-2947
- Acta Microbiologica Sinica, 63(8): 2935-2947
-
文章历史
- 收稿日期:2022-11-21
- 网络出版日期:2023-03-10
2. 河北省临床检验中心, 河北 石家庄 050000
2. Hebei Provincial Center for Clinical Laboratories, Shijiazhuang 050000, Hebei, China
艰难拟梭菌(Clostridioides difficile)是一种产芽孢的革兰阳性专性厌氧杆菌,通常存在于土壤、动物和人类肠道中,最初于1935年从健康新生儿粪便中分离得到。因其极度厌氧并且分离培养困难,最初依据表型特征将其归入梭菌属并命名为艰难梭菌(Clostridium difficile)[1]。2016年Lawson等[2]根据16S rRNA基因序列的特点提出将艰难梭菌归入消化链球菌科下的拟梭菌属(Clostridioides)。2019年美国微生物学会将“Clostridium difficile”正式更正为“Clostridioides difficile”,中文译名从“艰难梭菌”修改为“艰难拟梭菌”。艰难拟梭菌在不利于其生长的环境中会形成芽孢,芽孢抵抗力极强,能耐受高温和酒精,被宿主排出体外后可在外界环境中存活数月,并经粪口途径进一步传播[3-5]。为此,由艰难拟梭菌引起的艰难拟梭菌感染(Clostridioides difficile infection, CDI)是最常见的医院获得性感染之一,也是抗生素相关性腹泻(antibiotic-associated diarrhea, AAD)最常见的病因[4, 6]。CDI临床表现从轻度自限性腹泻到伪膜性结肠炎以及中毒性巨结肠和肠穿孔,严重可导致患者死亡[7]。目前公认的CDI危险因素包括:长期使用广谱抗生素、住院史和高龄(年龄≥65岁),其他危险因素还包括使用质子泵抑制剂、手术史,以及患有慢性基础疾病如消化道疾病等[8]。
根据艰难拟梭菌的产毒情况可将其分为产毒型和非产毒型菌株,产毒型艰难拟梭菌产生的毒素A (TcdA)和毒素B (TcdB)可失活宿主细胞GTP酶进而导致细胞骨架解聚和细胞坏死,在CDI过程中发挥关键作用[9-10]。除TcdA和TcdB之外,一些高产毒株还可分泌细胞致死性肿胀毒素(cytolethal distending toxin, CDT),即二元毒素,一种可破坏肌动蛋白细胞骨架的腺苷二磷酸(adenosine diphosphate, ADP)-核糖基转移酶[11]。除了引起肠上皮细胞坏死外,艰难拟梭菌毒素还能增加上皮通透性进而导致肠上皮水肿;破坏肠上皮细胞间紧密连接进而促进细菌移位至肠外组织;诱导免疫细胞分泌促炎因子如肿瘤坏死因子;募集中性粒细胞至炎症部位进而激发炎症反应[12]。Drewes等[13]研究发现,亚洲地区分离率较高的tcdA-tcdB+菌株同tcdA+ tcdB+菌株一样具有致病性,在患者和动物模型中均可导致感染,因此TcdB被认为是艰难拟梭菌感染中更关键的毒素。此外,研究人员还发现,艰难拟梭菌作为致癌细菌诱导结直肠肿瘤的发生发展及致瘤黏膜免疫反应,这一过程需要TcdB的参与[13]。鉴于毒素重要的病理生理学功能,本文将对艰难拟梭菌毒素的致病过程和影响毒素基因表达的分子调控机制进行综述。
1 TcdA和TcdB的结构域及其作用机制早期对毒素致病性的研究主要基于动物实验,Lyerly等[14]在仓鼠和小鼠模型中发现纯化的TcdA灌胃会诱导肠液积聚、肠道炎症反应、腹泻、出血和死亡,TcdB灌胃则无此现象,因此TcdA被称为肠毒素。当时普遍认为TcdA在CDI中发挥更重要的作用,TcdB可能辅助TcdA引发疾病。然而,后期研究表明TcdB比TcdA更具“杀伤力”,TcdB对结肠组织的破坏能力是TcdA的10倍,对HT29细胞的毒性比TcdA强数百倍,故TcdB又被称为细胞毒素[15-16]。TcdA和TcdB均存在4个功能结构域共同介导毒素发挥作用,包括N-末端葡萄糖基转移酶结合域(glucosyltransferase domain, GTD)、半胱氨酸蛋白酶结合域(cysteine protease domain, CPD)、易位域(transmembrane domain, TMD)和受体结合域(receptor-binding domain, RBD),RBD位于毒素蛋白的C末端,由重复的多肽单元组成,又称组合重复寡肽(combined repetitive oligopeptide, CROP)[17-18](图 1A)。4个功能结构域可以划分为A、B两个亚基,A亚基即GTD,能够通过糖基化作用使宿主细胞GTP酶失活,属于艰难拟梭菌毒素的毒性区。TcdA和TcdB的GTD序列相似性达74%,但TcdB的GTD活性更高,导致TcdB毒性是TcdA的100–1 000倍。B亚基由RBD、TMD和CPD构成,负责将A亚基转运到靶细胞中。RBD介导毒素与靶细胞表面受体结合后,TMD能够感应内体pH变化从而诱导毒素构象改变进而暴露疏水结构域,随后,CPD与宿主细胞衍生的分子六磷酸肌醇(inositol hexakisphosphate, InsP6)结合从而诱导GTD从内体释放到宿主细胞胞浆中发挥毒性作用(图 1C),下面将重点阐述毒素的作用机制。
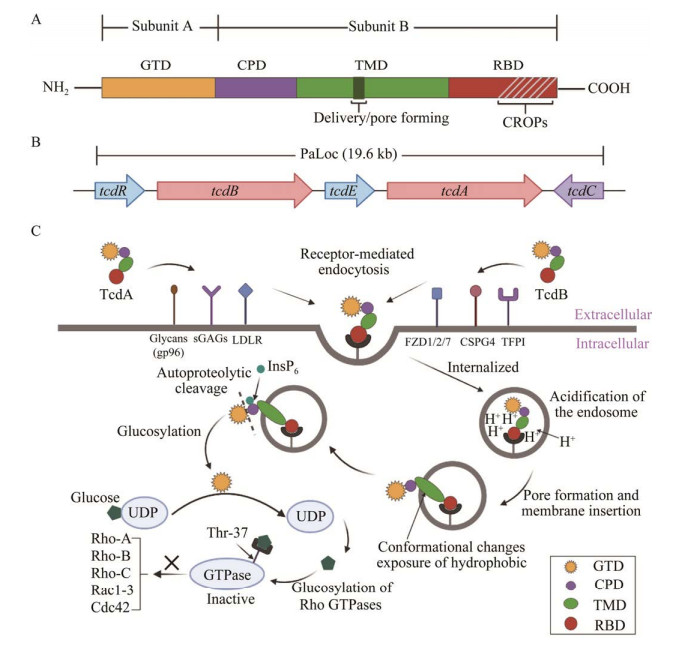
|
| 图 1 艰难拟梭菌TcdA和TcdB的结构及作用机制 Figure 1 Structure and mechanism of toxin A and toxin B in Clostridioides difficile. A: Four protein domain structures of toxin A and toxin B. B: Genetic arrangement of toxin A and toxin B pathogenicity locus. C: Molecular mechanism of toxins action. GTD: Glucosyltransferase domain; CPD: Cysteine protease domain; TMD: Transmembrane domain; RBD: Receptor-binding domain; CROPs: Combined repetitive oligopeptide. |
1.1 受体介导的胞吞作用
艰难拟梭菌在宿主体内定殖后产生毒素,毒素RBD与宿主细胞膜表面受体结合,通过内吞作用进入宿主细胞并以内体形式存在。研究发现细胞膜表面人糖蛋白受体gp96能够结合TcdA的RBD并介导毒素胞吞;而硫酸化糖胺聚糖(sulfated glycosaminoglycans, sGAGs)和低密度脂蛋白受体(low-density lipoprotein receptor, LDLR) 2种受体可直接介导TcdA的结合和进入而不依赖与RBD的结合[19]。TcdB有8种亚型,其中与人类CDI相关的主要是1–4型,研究证实卷曲蛋白(frizzled, FZD)成员1、2和7,以及硫酸软骨素蛋白多糖4 (chondroitin sulfate proteoglycan 4, CSPG4)是1型(TcdB1)和3型(TcdB3)结合和进入宿主细胞的高亲和力受体,而TcdB2和TcdB4主要与结肠中高度表达的组织因子途径抑制物(tissue factor pathway inhibit, TFPI)结合[20]。
1.2 内体酸化及毒性片段释放内体内环境酸化后引起毒素TMD构象改变,导致毒素蛋白的结构重排,位于TMD中的疏水区域暴露并形成突起,促使内体膜上形成孔道,为后续释放GTD提供条件[21]。研究表明TcdA的释放需要胆固醇协助形成孔道,而TcdB不需要[22]。毒素插入到内体膜上后,CPD与宿主细胞衍生的分子InsP6结合后促使毒素构象改变,A亚基与B亚基发生自催化切割,活性A亚基离开内体后进入宿主细胞浆内,B亚基仍留在内体膜上[23-24]。
1.3 糖基化作用使宿主细胞GTP酶失活Rho家族的GTP酶能够将GTP水解成GDP,在调节细胞周期、介导细胞间,以及细胞与上皮间的黏附、维持细胞骨架完整性等方面发挥重要作用。活性GTD释放到宿主胞浆中后,以UDP-葡萄糖为共底物,将自身糖基转移到GTP酶的Thr-37上从而使Rho家族(Rho-A, -B, and -C)、Rac1-3和Cdc42等小GTP酶失活,引起肌动蛋白解聚,导致细胞骨架重新排列,影响细胞结构完整性,最终引起靶细胞的凋亡。随后,细胞间紧密连接被破坏,肠上皮通透性增加,引起肠道水肿等病理改变[4]。
2 tcdA和tcdB的表达与调控多种调节因子通过直接或间接方式调控tcdA和tcdB的表达,进而影响艰难拟梭菌产生毒素。下文简要介绍3种毒素致病基因座内的调节基因和6种致病基因座外的调节因子。
2.1 毒素致病基因座内的调节产毒型艰难拟梭菌的毒素基因tcd位于一个19.6 kb的致病基因座(pathogenicity locus, PaLoc)内,PaLoc阅读框内包含tcdA和tcdB 2个毒素基因,以及tcdR、tcdE和tcdC 3个毒素调节基因(图 1B)。tcdR和tcdE被激活后可促进tcdA和tcdB的表达,而tcdC作为负向调控基因,对tcdA和tcdB表达主要起抑制作用。
2.1.1 TcdRTcdR是一种对毒素基因表达至关重要的一种蛋白,通常在艰难拟梭菌生长进入平台期后活性增强,通过招募RNA聚合酶到毒素基因和自身的启动子,正向调控tcdA、tcdB和自身的转录[25]。一项针对艰难拟梭菌R20291菌株的研究显示,TcdR除参与毒素产生外,还影响芽孢形成,tcdR基因突变型菌株的芽孢形成率和萌发率均低于野生型菌株[26]。前期研究中,发现tcdR基因在不同菌株中表达情况不同。根据tcdR基因在不同菌株中的存在与否,本研究将其作为一个靶位点,建立了艰难拟梭菌二元基因分型方法[27]。
2.1.2 TcdETcdE与Holin家族蛋白具有同源性,其编码基因tcdE在PaLoc上位于tcdA和tcdB之间。Holin家族蛋白是一种疏水性跨膜蛋白,能够插入胞膜形成非特异性通道,介导胞内外物质的跨膜转运。TcdE或可通过Holin跨膜转运系统的方式促进艰难拟梭菌毒素向细胞外环境的释放[28]。
2.1.3 TcdCTcdC是一种在艰难拟梭菌生长指数期高表达的酸性膜相关蛋白,通过干扰TcdR与RNA聚合酶的识别,负向调控艰难拟梭菌毒素的生成[29]。研究发现核糖体分型(ribotyping, RT) 027型高产毒艰难拟梭菌的tcdC基因117位单碱基对移码突变或18 bp片段缺失,造成tcdC基因功能丧失[30]。Carter等[31]用携带了tcdC质粒的RT027型菌株M7404感染仓鼠,发现重组菌株的产毒能力和致病性均降低,说明tcdC基因突变丧失了对毒素的负调控作用,可能与RT027型艰难拟梭菌的毒素产量增加有关。但其他研究产生了相反的结论,Cartman等[32]在研究中发现tcdC基因型和毒素产量之间无任何联系。通过比较低产毒菌株CD630和高产毒菌株RT027型的毒素产生水平,Anwar等[33]同样发现RT027型菌株之间产毒水平存在很大差异,分泌的总毒素量等于甚至低于CD630。上述研究表明,tcdC基因的功能及其在致病过程中所起的作用仍需要进行更加深入的研究。
在非产毒型菌株中,PaLoc被75–115个核苷酸非编码序列或7.2 kb功能未知序列所取代,有研究表明非产毒艰难拟梭菌菌株CCUG37785定殖于肠道可预防产毒株的定殖,显著降低CDI的复发率[34]。然而,Brouwer等[35]发现非产毒艰难拟梭菌可以通过水平基因转移从产毒型菌株中获得PaLoc,进而转化为产毒型菌株诱导宿主肠道炎症反应。鉴于非产毒艰难拟梭菌在CDI中的作用存在较大的科学争论,未来仍需对非产毒型菌株进行深入研究。
2.2 致病基因座外的调节因子 2.2.1 芽孢形成转录因子芽孢形成转录因子(stage 0 sporulation protein A, Spo0A)是一种普遍存在于产芽孢病原菌中的DNA结合蛋白,参与细菌毒素产生、芽孢萌发、鞭毛表达和生物膜形成等多项生理活动。近年来通过Clos Tron技术来构建艰难拟梭菌基因敲除模型,加快了对艰难拟梭菌spo0A基因的研究。通过比较∆spo0A突变株与亲代株间TcdA和TcdB表达差异和致病基因座上相关基因转录水平,Song等[36]发现Spo0A负向调控毒素产生。相比亲代菌株C25 (RT014型),C25 ∆spo0A毒素产量增多,其中TcdA蛋白含量更为显著。同时,突变株中tcdA、tcdB、tcdE和tcdR基因表达水平均高于亲代株。Pettit等[37]观察到CD630 ∆spo0A突变株的TcdA生成量显著高于野生型菌株。这与Underwood等[38]的研究结果形成鲜明对比,他们发现CD630 ∆erm spo0A突变株中TcdA的表达水平远低于亲代株,体外细胞实验同样显示spo0A突变株对Vero细胞毒性作用显著下降。可见,spo0A基因对艰难拟梭菌毒素基因的调节作用目前仍存在较大争议,spo0A对艰难拟梭菌毒素产生的影响及机制还有待进一步研究。
2.2.2 RstA蛋白RstA是多功能RGG/Rap/NprR/PlcR/PRGX (RRNPP)家族的成员,能够调控艰难拟梭菌毒素产生和芽孢形成[39]。Edwards等[40]观察到RstA蛋白能够与tcdR、tcdA和tcdB基因的启动子区域直接结合来抑制毒素基因和毒素调控基因的转录,并通过影响sigD转录从而间接抑制毒素的产生。此外,通过构建R20291 ∆rstA突变株,Edwards等[41]同样证明相比于低产毒株CD630 Δerm,RstA蛋白对高产毒RT027型菌株产毒抑制作用更大。
2.2.3 全局性转录调控因子全局性转录调控因子(GTP-sensing transcriptional pleiotropic repressor, CodY)普遍存在于低G+C含量的革兰阳性菌中,与支链氨基酸和/或GTP结合后被激活,在调控细菌毒力表达方面发挥关键作用。例如,在单核细胞增多性李斯特菌(Listeria monocytogenes)中,CodY可直接与毒力激活基因prfA结合进而促进细菌产毒[42]。相反,在艰难拟梭菌中,CodY与RNA聚合酶竞争结合TcdR启动子区域,干扰TcdR的正常功能从而间接抑制艰难拟梭菌毒素的产生。此外,Edwards等[41]表明CodY抑制tcdR基因的能力在不同型别菌株中存在差异,其中对CD630 ∆erm菌株的产毒抑制作用最强。
2.2.4 分解代谢控制蛋白A分解代谢控制蛋白A (catabolite control protein A, CcpA)是一种多功能调节子,参与糖摄取、糖酵解和氨基酸代谢等许多代谢途径。早期研究发现葡萄糖可抑制艰难拟梭菌毒素基因的表达[43],Antunes等[44]进一步证实这一过程受到CcpA的调节,他们发现在ccpA失活的艰难拟梭菌中,葡萄糖对毒素抑制作用也随之减弱。
2.2.5 σ因子σ因子是RNA聚合酶的辅助因子,与RNA聚合酶结合后转变为聚合酶全酶。研究发现,艰难拟梭菌CD630 ∆sigH菌株的tcdA、tcdB和tcdR表达均上调,这表明SigH对艰难拟梭菌毒素基因起负调控作用[45]。另一种σ因子SigD能够直接激活tcdR进而间接提高tcdA和tcdB基因表达水平,这个过程受到第二信使环二鸟苷酸(cyclic diguanylate, c-di-GMP)的调控,菌体内c-di-GMP浓度的升高可显著降低sigD mRNA水平,sigD对tcdR的激活作用也随之减弱,进而间接抑制tcdA和tcdB的表达[46]。
2.2.6 Sporulation inhibitor (sin)位点Sporulation inhibitor (sin)位点是一个双基因操纵子,艰难拟梭菌的基因组中有2个sinR的同源基因sinR和sinR',二者互为拮抗作用。SinR是芽孢形成、毒素产生和运动能力的正向调节因子,SinR′则通过抑制SinR的活性从而抑制毒素的产生。研究发现敲除sinRR'基因的菌株中c-di-GMP浓度比野生型菌株增加了3倍,因此sinRR'突变株可能通过增加c-di-GMP的浓度来抑制sigD表达进而导致艰难拟梭菌的毒素产量显著减少[47-49]。
2.2.7 鞭毛蛋白基因鞭毛在艰难拟梭菌对宿主肠道的定殖中至关重要,而近年来的研究更多关注了鞭毛蛋白基因对艰难拟梭菌毒力的调节。鞭毛蛋白基因众多且不同基因对艰难拟梭菌毒力的影响不尽相同,研究发现fliM、fliF、fliG和flhB-fliR均对毒素基因表达起促进作用,相反,fliC和fliD能抑制毒素基因的表达[50-51]。多种鞭毛蛋白基因是如何协调对艰难拟梭菌毒素基因表达的影响尚不明确,更深层的调节机制仍有待探索。
总之,艰难拟梭菌毒素的表达与调控是一个复杂且精细的过程,致病基因座内外的多种调节因子是如何协同调节菌体在不同生长周期和生存环境中的毒素表达有待深入探讨。
3 CDT的结构域及其作用机制CDT同样由2种组分构成:活化ADP核糖基转移酶的酶组分(CDTa)和协助酶组分进入细胞溶质的结合/易位组分(CDTb,图 2A)。CDT被丝氨酸样蛋白激活后,CDTb相继与宿主细胞膜上的脂蛋白受体(lipoprotein receptor, LSR)及CDTa结合形成复合物并以内吞的形式进入靶细胞形成内体[52]。随后,内体酸化引起CDTb插入内体膜,形成孔道介导CDTa释放到宿主细胞胞浆中,CDTa通过抑制G-肌动蛋白聚合和促进F-肌动蛋白解聚导致肌动蛋白细胞骨架的完全破坏,最终导致细胞死亡(图 2C)[53]。此外,CDT还影响细胞蛋白的磷酸化状态,最近的研究表明,用CDT处理Hep-2细胞后,近1 100个磷酸化位点的磷酸化水平发生变化[54]。研究发现,CDT在小鼠体内主要以Toll样受体2 (Toll-like receptor 2, TLR2)依赖的方式抑制炎症状态下嗜酸性粒细胞对结肠的保护作用,进而加重肠上皮组织炎症。与感染野生型高产毒艰难拟梭菌R20291的小鼠相比,感染R20291 ∆cdtB菌株的小鼠肠道内的嗜酸性粒细胞明显增多,结肠的炎症也明显减轻,这进一步证实了CDT在艰难拟梭菌致病过程中的重要作用[55]。除结肠嗜酸性粒细胞外,CDT还可通过激活上皮和黏膜等部位的黏膜相关恒定T细胞(mucosal-associated invariant T, MAIT)发挥毒性作用。MAIT受CDT激活后可诱导快速的固有免疫反应,产生细胞毒性物质如穿孔素和颗粒酶B,导致肠上皮细胞的损伤和上皮屏障的破坏[56]。即使在亚致死毒素浓度下,CDT也可以诱导上皮屏障功能障碍。
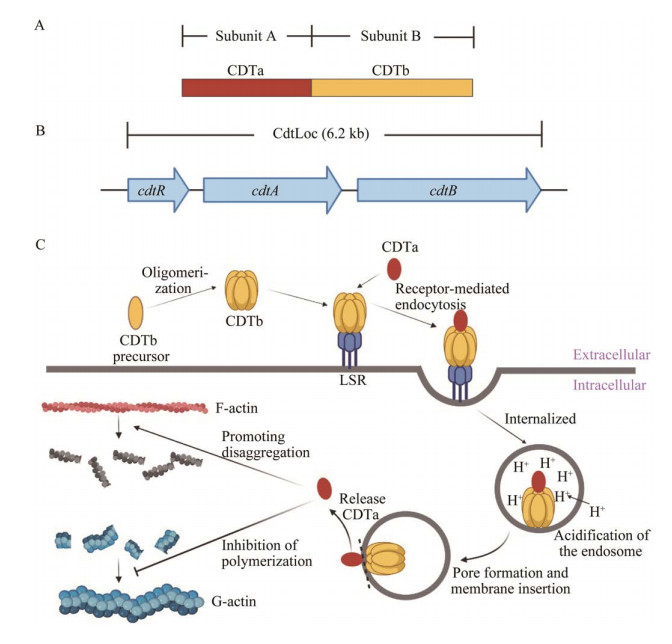
|
| 图 2 艰难拟梭菌CDT的结构及作用机制 Figure 2 Structure and mechanism of binary toxin in Clostridioides difficile. A: Protein domain structures of binary toxin. B: Genetic arrangement of binary toxin CDT locus. C: Molecular mechanism of binary toxin action. |
4 cdt的表达与调控
近年来,027/NAP1/BI (核糖体分型:027,脉冲场凝胶电泳分型:NAP1,限制性内切酶分型:BI)高产毒艰难拟梭菌在欧美国家多次暴发流行,高致病性和高死亡率使其迅速成为全球医疗卫生系统的焦点[57]。027/NAP1/BI最显著的毒素表型之一就是产生CDT。二元毒素基因cdt位于一个6.2 kb的致病基因座(CDT locus, CDTLoc)内,内含cdtA、cdtB和cdtR 3个基因[58]。CdtR不仅是cdtA和cdtB表达的正向调控因子,还可促进TcdA和TcdB的产生(图 2B)[59]。临床证据表明,cdtB基因阳性菌株感染与重症密切相关,会增加疾病严重程度、感染复发和死亡的风险[60]。此外,二元毒素基因阳性的艰难拟梭菌对抗生素的敏感性降低,一些菌株如RT027、RT018和RT356表现出了多重耐药性[61]。
5 问题与展望影响毒素产生的基因繁多且调控过程复杂,难以依据一种调控基因对产毒水平进行定性和定量分析。除上述调控基因外,菌体内是否存在其他可影响产毒的分子,多种调控因子共同作用下毒素的表达如何平衡以及如何针对调控基因来减少产毒等问题仍有待进一步的研究。毒素作为艰难拟梭菌最主要的毒力因子,其致病机制和调控因子的阐明为研制新型治疗或预防药物提供了新思路。针对艰难拟梭菌毒素开发的新药物如单克隆抗体和小分子抑制剂也越来越多。Bezlotoxumab单抗通过识别TcdB C-末端CROP从而中和毒素,是唯一被美国食品药品监督管理局(Food and Drug Administration, FDA)批准应用于复发CDI风险的患者的辅助治疗药物。类似地,以TcdA和TcdB的GTD结构域或InsP6为靶点研制的小分子抑制剂,其治疗效果在体外试验或动物实验中均得以验证[24, 62]。总之,作用于艰难拟梭菌毒素尤其是其调控基因的治疗药物仍十分有限,现有新药物的安全性和有效性仍需进一步研究。
许多抗生素,尤其是广谱抗生素,对靶细菌的杀灭作用不存在特异性,可能影响正常肠道菌群。艰难拟梭菌毒素的蛋白结构或表面相关分子可能同样存在于肠道共生微生物中,因此对于抗毒素小分子抑制剂的研制应更多的关注TcdA和TcdB的特异性位点,以免破坏正常微生物群的动态平衡。另外,最新研究表明毒素产量和毒力之间并不是单纯的正相关,高产毒RT027型菌株之间毒素产量存在很大差异,其中有些甚至低于一般产毒株。虽然TcdA和TcdB作为艰难拟梭菌感染的关键毒力因子已达成共识,但无证据表明毒素量需达到一定阈值才会引起相关的临床症状。因此,感染菌株产毒水平与疾病严重程度之间的关系还有待进一步探讨,对于出现腹泻等症状但粪便中未检测到毒素的患者同样应予以重视。RT027型艰难拟梭菌兼具高产毒和高耐药的特点,其不仅产生二元毒素,还对氟喹诺酮类抗生素耐药,二元毒素表达与RT027型艰难拟梭菌高耐药表型之间的相关性值得深入研究。在艰难拟梭菌培养的基础上,对临床分离的菌株进行分子分型、毒素基因检测和耐药分析将是一种更加适合临床的个性化治疗策略。
总之,对艰难拟梭菌毒素表达和致病机制的研究还应更深入,这对开发针对这些机制的有效治疗策略,实现CDI的有效防治具有重要意义。
| [1] | BURNHAM CAD, CARROLL KC. Diagnosis of Clostridium difficile infection: an ongoing conundrum for clinicians and for clinical laboratories[J]. Clinical Microbiology Reviews, 2013, 26(3): 604-630 DOI:10.1128/CMR.00016-13. |
| [2] | LAWSON PA, CITRON DM, TYRRELL KL, FINEGOLD SM. Reclassification of Clostridium difficile as Clostridioides difficile (Hall and O'Toole 1935) Prévot 1938[J]. Anaerobe, 2016, 40: 95-99 DOI:10.1016/j.anaerobe.2016.06.008. |
| [3] | ZHU DL, SORG JA, SUN XM. Clostridioides difficile biology: sporulation, germination, and corresponding therapies for C. difficile infection[J]. Frontiers in Cellular and Infection Microbiology, 2018, 8: 29 DOI:10.3389/fcimb.2018.00029. |
| [4] | ABT MC, MCKENNEY PT, PAMER EG. Clostridium difficile colitis: pathogenesis and host defence[J]. Nature Reviews Microbiology, 2016, 14(10): 609-620 DOI:10.1038/nrmicro.2016.108. |
| [5] | MONTES-BRAVO N, ROMERO-RODRÍGUEZ A, GARCÍA-YUNGE J, MEDINA C, PIZARRO-GUAJARDO M, PAREDES-SABJA D. Role of the spore coat proteins CotA and CotB, and the spore surface protein CDIF630_02480, on the surface distribution of exosporium proteins in Clostridioides difficile 630 spores[J]. Microorganisms, 2022, 10(10): 1918 DOI:10.3390/microorganisms10101918. |
| [6] | CURCIO D, CANÉ A, FERNÁNDEZ FA, CORREA J. Clostridium difficile-associated diarrhea in developing countries: a systematic review and meta-analysis[J]. Infectious Diseases and Therapy, 2019, 8(1): 87-103 DOI:10.1007/s40121-019-0231-8. |
| [7] | TIAN SH, XIONG XZ, ZENG J, WANG SY, TREMBLAY BJM, CHEN P, CHEN BH, LIU M, CHEN PS, SHENG KW, ZEVE D, QI WS, BREAULT DT, RODRÍGUEZ C, GERHARD R, JIN RS, DOXEY AC, DONG M. Identification of TFPI as a receptor reveals recombination-driven receptor switching in Clostridioides difficile toxin B variants[J]. Nature Communications, 2022, 13: 6786 DOI:10.1038/s41467-022-33964-9. |
| [8] | ZHANG J, CHEN L, GOMEZ-SIMMONDS A, YIN MT, FREEDBERG DE. Antibiotic-specific risk for community-acquired Clostridioides difficile infection in the United States from 2008 to 2020[J]. Antimicrobial Agents and Chemotherapy, 2022, 66(12): e0112922 DOI:10.1128/aac.01129-22. |
| [9] | ORRELL KE, MELNYK RA. Large clostridial toxins: mechanisms and roles in disease[J]. Microbiology and Molecular Biology Reviews: MMBR, 2021, 85(3): e0006421 DOI:10.1128/MMBR.00064-21. |
| [10] | PRUSS KM, SONNENBURG JL. C. difficile exploits a host metabolite produced during toxin-mediated disease[J]. Nature, 2021, 593(7858): 261-265 DOI:10.1038/s41586-021-03502-6. |
| [11] | KAWAMOTO A, YAMADA T, YOSHIDA T, SATO Y, KATO T, TSUGE H. Cryo-EM structures of the translocational binary toxin complex CDTa-bound CDTb-pore from Clostridioides difficile[J]. Nature Communications, 2022, 13: 6119 DOI:10.1038/s41467-022-33888-4. |
| [12] | LI Y, XU S, XU QQ, CHEN YJ. Clostridium difficile toxin B induces colonic inflammation through the TRIM46/DUSP1/MAPKs and NF-κB signalling pathway[J]. Artificial Cells, Nanomedicine, and Biotechnology, 2020, 48(1): 452-462 DOI:10.1080/21691401.2019.1709856. |
| [13] | DREWES JL, CHEN J, MARKHAM NO, KNIPPEL RJ, DOMINGUE JC, TAM AJ, CHAN JL, KIM L, McMANN M, STEVENS C, DEJEA CM, TOMKOVICH S, MICHEL J, WHITE JR, MOHAMMAD F, CAMPODÓNICO VL, HEISER CN, WU XQ, WU SG, DING H, et al. Human colon cancer-derived Clostridioides difficile strains drive colonic tumorigenesis in mice[J]. Cancer Discovery, 2022, 12(8): 1873-1885 DOI:10.1158/2159-8290.CD-21-1273. |
| [14] | LYERLY DM, SAUM KE, MacDONALD DK, WILKINS TD. Effects of Clostridium difficile toxins given intragastrically to animals[J]. Infection and Immunity, 1985, 47(2): 349-352 DOI:10.1128/iai.47.2.349-352.1985. |
| [15] | CARTER GP, CHAKRAVORTY A, PHAM NGUYEN TA, MILETO S, SCHREIBER F, LI L, HOWARTH P, CLARE S, CUNNINGHAM B, SAMBOL SP, CHEKNIS A, FIGUEROA I, JOHNSON S, GERDING D, ROOD JI, DOUGAN G, LAWLEY TD, LYRAS D. Defining the roles of TcdA and TcdB in localized gastrointestinal disease, systemic organ damage, and the host response during Clostridium difficile infections[J]. mBio, 2015, 6(3): e00551. |
| [16] | LI ZH, LEE K, RAJYAGURU U, JONES CH, JANEZIC S, RUPNIK M, ANDERSON AS, LIBERATOR P. Ribotype classification of Clostridioides difficile isolates is not predictive of the amino acid sequence diversity of the toxin virulence factors TcdA and TcdB[J]. Frontiers in Microbiology, 2020, 11: 1310 DOI:10.3389/fmicb.2020.01310. |
| [17] | JIANG MQ, SHIN J, SIMEON R, CHANG JY, MENG R, WANG YH, SHINDE O, LI PW, CHEN ZL, ZHANG JJ. Structural dynamics of receptor recognition and pH-induced dissociation of full-length Clostridioides difficile toxin B[J]. PLoS Biology, 2022, 20(3): e3001589 DOI:10.1371/journal.pbio.3001589. |
| [18] | AMINZADEH A, LARSEN CE, BOESEN T, JØRGENSEN R. High-resolution structure of native toxin A from Clostridioides difficile[J]. EMBO Reports, 2022, 23(1): e53597 DOI:10.15252/embr.202153597. |
| [19] | TAO L, TIAN SH, ZHANG J, LIU ZM, ROBINSON-MCCARTHY L, MIYASHITA SI, BREAULT DT, GERHARD R, OOTTAMASATHIEN S, WHELAN SPJ, DONG M. Sulfated glycosaminoglycans and low-density lipoprotein receptor contribute to Clostridium difficile toxin A entry into cells[J]. Nature Microbiology, 2019, 4(10): 1760-1769 DOI:10.1038/s41564-019-0464-z. |
| [20] | LUO JH, YANG Q, ZHANG XF, ZHANG YY, WAN L, ZHAN XC, ZHOU Y, HE LQ, LI DY, JIN DZ, ZHEN Y, HUANG J, LI YY, TAO L. TFPI is a colonic crypt receptor for TcdB from hypervirulent clade 2 C. difficile[J]. Cell, 2022, 185(6): 980-994.e15 DOI:10.1016/j.cell.2022.02.010. |
| [21] | ORRELL KE, TELLGREN-ROTH Å, di BERNARDO M, ZHANG ZF, CUVIELLO F, LUNDQVIST J, von HEIJNE G, NILSSON I, MELNYK RA. Direct detection of membrane-inserting fragments defines the translocation pores of a family of pathogenic toxins[J]. Journal of Molecular Biology, 2018, 430(18 Pt B): 3190-3199. |
| [22] | PAPATHEODOROU P, SONG S, LÓPEZ-UREÑA D, WITTE A, MARQUES F, OST GS, SCHORCH B, CHAVES-OLARTE E, AKTORIES K. Cytotoxicity of Clostridium difficile toxins A and B requires an active and functional SREBP-2 pathway[J]. FASEB Journal: Official Publication of the Federation of American Societies for Experimental Biology, 2019, 33(4): 4883-4892 DOI:10.1096/fj.201801440R. |
| [23] | BILVERSTONE TW, GARLAND M, CAVE RJ, KELLY ML, THOLEN M, BOULEY DM, KAYE P, MINTON NP, BOGYO M, KUEHNE SA, MELNYK RA. The glucosyltransferase activity of C. difficile toxin B is required for disease pathogenesis[J]. PLoS Pathogens, 2020, 16(9): e1008852 DOI:10.1371/journal.ppat.1008852. |
| [24] | IVARSSON ME, DURANTIE E, HUBERLI C, HUWILER S, HEGDE C, FRIEDMAN J, ALTAMURA F, LU J, VERDU EF, BERCIK P, LOGAN SM, CHEN WX, LEROUX JC, CASTAGNER B. Small-molecule allosteric triggers of Clostridium difficile toxin B auto-proteolysis as a therapeutic strategy[J]. Cell Chemical Biology, 2019, 26(1): 17-26.e13 DOI:10.1016/j.chembiol.2018.10.002. |
| [25] | XU XX, LUO Y, CHEN H, SONG XJ, BIAN Q, WANG XJ, LIANG Q, ZHAO JH, LI CH, SONG GZ, YANG J, SUN LL, JIANG JM, WANG HY, ZHU B, YE GY, CHEN L, TANG YW, JIN DZ. Genomic evolution and virulence association of Clostridioides difficile sequence type 37 (ribotype 017) in China[J]. Emerging Microbes & Infections, 2021, 10(1): 1331-1345. |
| [26] | GIRINATHAN BP, MONOT M, BOYLE D, McALLISTER KN, SORG JA, DUPUY B, GOVIND R. Effect of tcdR mutation on sporulation in the epidemic Clostridium difficile strain R20291[J]. mSphere, 2017, 2(1): e00383-e00316. |
| [27] | LI ZR, LIU XL, ZHAO JH, XU KY, TIAN TT, YANG J, QIANG CX, SHI DY, WEI HL, SUN SJ, CUI QQ, LI RX, NIU YN, HUANG BX. Comparison of a newly developed binary typing with ribotyping and multilocus sequence typing methods for Clostridium difficile[J]. Journal of Microbiological Methods, 2018, 147: 50-55 DOI:10.1016/j.mimet.2018.02.012. |
| [28] | VIDOR CJ, HAMIOT A, WISNIEWSKI J, MATHIAS RA, DUPUY B, AWAD M, LYRAS D. A highly specific holin-mediated mechanism facilitates the secretion of lethal toxin TcsL in Paeniclostridiumsordellii[J]. Toxins, 2022, 14(2): 124 DOI:10.3390/toxins14020124. |
| [29] | DINGLE KE, ELLIOTT B, ROBINSON E, GRIFFITHS D, EYRE DW, STOESSER N, VAUGHAN A, GOLUBCHIK T, FAWLEY WN, WILCOX MH, PETO TE, WALKER AS, RILEY TV, CROOK DW, DIDELOT X. Evolutionary history of the Clostridium difficile pathogenicity locus[J]. Genome Biology and Evolution, 2014, 6(1): 36-52 DOI:10.1093/gbe/evt204. |
| [30] | JAZMATI N, HAIN O, HELLMICH M, PLUM G, KAASCH A. PCR based detection of tcdCΔ117 in Clostridium difficile infection identifies patients at risk for recurrence-a hospital-based prospective observational study[J]. Anaerobe, 2019, 57: 39-44 DOI:10.1016/j.anaerobe.2019.03.010. |
| [31] | CARTER GP, DOUCE GR, GOVIND R, HOWARTH PM, MACKIN KE, SPENCER J, BUCKLEY AM, ANTUNES A, KOTSANAS D, JENKIN GA, DUPUY B, ROOD JI, LYRAS D. The anti-sigma factor TcdC modulates hypervirulence in an epidemic BI/NAP1/027 clinical isolate of Clostridium difficile[J]. PLoS Pathogens, 2011, 7(10): e1002317 DOI:10.1371/journal.ppat.1002317. |
| [32] | CARTMAN ST, KELLY ML, HEEG D, HEAP JT, MINTON NP. Precise manipulation of the Clostridium difficile chromosome reveals a lack of association between the tcdC genotype and toxin production[J]. Applied and Environmental Microbiology, 2012, 78(13): 4683-4690 DOI:10.1128/AEM.00249-12. |
| [33] | ANWAR F, ROXAS BAP, SHEHAB KW, AMPEL NM, VISWANATHAN VK, VEDANTAM G. Low-toxin Clostridioides difficile RT027 strains exhibit robust virulence[J]. Emerging Microbes & Infections, 2022, 11(1): 1982-1993. |
| [34] | WANG SH, HEULER J, WICKRAMAGE I, SUN XM. Genomic and phenotypic characterization of the nontoxigenic Clostridioides difficile strain CCUG37785 and demonstration of its therapeutic potential for the prevention of C. difficile infection[J]. Microbiology Spectrum, 2022, 10(2): e0178821 DOI:10.1128/spectrum.01788-21. |
| [35] | BROUWER MSM, ROBERTS AP, HUSSAIN H, WILLIAMS RJ, ALLAN E, MULLANY P. Horizontal gene transfer converts non-toxigenic Clostridium difficile strains into toxin producers[J]. Nature Communications, 2013, 4: 2601 DOI:10.1038/ncomms3601. |
| [36] |
宋晓蕾, 周芬芬, 胥腾, 王莉, 李静文, 黄海辉. spo0A对艰难梭菌毒素A/B蛋白表达的调节作用[J]. 中国感染与化疗杂志, 2021, 21(2): 136-140.
SONG XL, ZHOU FF, XU T, WANG L, LI JW, HUANG HH. The regulatory role of spo0A in Clostridioides difficile toxin A and toxin B production[J]. Chinese Journal of Infection and Chemotherapy, 2021, 21(2): 136-140 (in Chinese). |
| [37] | PETTIT LJ, BROWNE HP, YU L, SMITS WK, FAGAN RP, BARQUIST L, MARTIN MJ, GOULDING D, DUNCAN SH, FLINT HJ, DOUGAN G, CHOUDHARY JS, LAWLEY TD. Functional genomics reveals that Clostridium difficile Spo0A coordinates sporulation, virulence and metabolism[J]. BMC Genomics, 2014, 15: 160 DOI:10.1186/1471-2164-15-160. |
| [38] | UNDERWOOD S, GUAN S, VIJAYASUBHASH V, BAINES SD, GRAHAM L, LEWIS RJ, WILCOX MH, STEPHENSON K. Characterization of the sporulation initiation pathway of Clostridium difficile and its role in toxin production[J]. Journal of Bacteriology, 2009, 191(23): 7296-7305 DOI:10.1128/JB.00882-09. |
| [39] | EDWARDS AN, ANJUWON-FOSTER BR, McBRIDE SM. RstA is a major regulator of Clostridioides difficile toxin production and motility[J]. mBio, 2019, 10(2): e01991-e01918. |
| [40] | EDWARDS AN, TAMAYO R, McBRIDE SM. A novel regulator controls Clostridium difficile sporulation, motility and toxin production[J]. Molecular Microbiology, 2016, 100(6): 954-971 DOI:10.1111/mmi.13361. |
| [41] | EDWARDS AN, KRALL EG, McBRIDE SM. Strain-dependent RstA regulation of Clostridioides difficile toxin production and sporulation[J]. Journal of Bacteriology, 2020, 202(2): e00586-e00519. |
| [42] | LOBEL L, SIGAL N, BOROVOK I, BELITSKY BR, SONENSHEIN AL, HERSKOVITS AA. The metabolic regulator CodY links Listeria monocytogenes metabolism to virulence by directly activating the virulence regulatory gene, prfA[J]. Molecular Microbiology, 2015, 95(4): 624-644 DOI:10.1111/mmi.12890. |
| [43] | BOUILLAUT L, DUBOIS T, SONENSHEIN AL, DUPUY B. Integration of metabolism and virulence in Clostridium difficile[J]. Research in Microbiology, 2015, 166(4): 375-383 DOI:10.1016/j.resmic.2014.10.002. |
| [44] | ANTUNES A, CAMIADE E, MONOT M, COURTOIS E, BARBUT F, SERNOVA NV, RODIONOV DA, MARTIN-VERSTRAETE I, DUPUY B. Global transcriptional control by glucose and carbon regulator CcpA in Clostridium difficile[J]. Nucleic Acids Research, 2012, 40(21): 10701-10718 DOI:10.1093/nar/gks864. |
| [45] | SAUJET L, MONOT M, DUPUY B, SOUTOURINA O, MARTIN-VERSTRAETE I. The key sigma factor of transition phase, SigH, controls sporulation, metabolism, and virulence factor expression in Clostridium difficile[J]. Journal of Bacteriology, 2011, 193(13): 3186-3196 DOI:10.1128/JB.00272-11. |
| [46] | MCKEE RW, MANGALEA MR, PURCELL EB, BORCHARDT EK, TAMAYO R. The second messenger cyclic Di-GMP regulates Clostridium difficile toxin production by controlling expression of sigD[J]. Journal of Bacteriology, 2013, 195(22): 5174-5185 DOI:10.1128/JB.00501-13. |
| [47] | CIFTCI Y, GIRINATHAN BP, DHUNGEL BA, HASAN MK, GOVIND R. Clostridioides difficile SinR' regulates toxin, sporulation and motility through protein-protein interaction with SinR[J]. Anaerobe, 2019, 59: 1-7 DOI:10.1016/j.anaerobe.2019.05.002. |
| [48] | GIRINATHAN BP, OU JJ, DUPUY B, GOVIND R. Pleiotropic roles of Clostridium difficile sin locus[J]. PLoS Pathogens, 2018, 14(3): e1006940 DOI:10.1371/journal.ppat.1006940. |
| [49] |
王伟刚, 杨靖, 牛亚楠, 赵建宏. 艰难类梭菌芽胞形成和萌发相关调控的研究进展[J]. 微生物学通报, 2021, 48(4): 1314-1322.
DOI:10.13344/j.microbiol.china.200596 WANG WG, YANG J, NIU YN, ZHAO JH. Advances in regulation of Clostridioides difficile sporulation and germination[J]. Microbiology China, 2021, 48(4): 1314-1322 (in Chinese). |
| [50] | AUBRY A, HUSSACK G, CHEN WX, KuoLEE R, TWINE SM, FULTON KM, FOOTE S, CARRILLO CD, TANHA J, LOGAN SM. Modulation of toxin production by the flagellar regulon in Clostridium difficile[J]. Infection and Immunity, 2012, 80(10): 3521-3532 DOI:10.1128/IAI.00224-12. |
| [51] | ZHU DL, WANG SH, SUN XM. FliW and CsrA govern flagellin (FliC) synthesis and play pleiotropic roles in virulence and physiology of Clostridioides difficile R20291[J]. Frontiers in Microbiology, 2021, 12: 735616 DOI:10.3389/fmicb.2021.735616. |
| [52] | HEMMASI S, CZULKIES BA, SCHORCH B, VEIT A, AKTORIES K, PAPATHEODOROU P. Interaction of the Clostridium difficile binary toxin CDT and its host cell receptor, lipolysis-stimulated lipoprotein receptor (LSR)[J]. The Journal of Biological Chemistry, 2015, 290(22): 14031-14044 DOI:10.1074/jbc.M115.650523. |
| [53] | GERDING DN, JOHNSON S, RUPNIK M, AKTORIES K. Clostridium difficile binary toxin CDT: mechanism, epidemiology, and potential clinical importance[J]. Gut Microbes, 2014, 5(1): 15-27 DOI:10.4161/gmic.26854. |
| [54] | STIEGLITZ F, GERHARD R, PICH A. The binary toxin of Clostridioides difficile alters the proteome and phosphoproteome of HEp-2 cells[J]. Frontiers in Microbiology, 2021, 12: 725612 DOI:10.3389/fmicb.2021.725612. |
| [55] | COWARDIN CA, BUONOMO EL, SALEH MM, WILSON MG, BURGESS SL, KUEHNE SA, SCHWAN C, EICHHOFF AM, KOCH-NOLTE F, LYRAS D, AKTORIES K, MINTON NP, PETRI WA. The binary toxin CDT enhances Clostridium difficile virulence by suppressing protective colonic eosinophilia[J]. Nature Microbiology, 2016, 1: 16108 DOI:10.1038/nmicrobiol.2016.108. |
| [56] | MARQUARDT I, JAKOB J, SCHEIBEL J, HOFMANN JD, KLAWONN F, NEUMANN-SCHAAL M, GERHARD R, BRUDER D, JÄNSCH L. Clostridioides difficile toxin CDT induces cytotoxic responses in human mucosal-associated invariant T (MAIT) cells[J]. Frontiers in Microbiology, 2021, 12: 752549 DOI:10.3389/fmicb.2021.752549. |
| [57] | FATIMA R, AZIZ M. The hypervirulent strain of Clostridium difficile: NAP1/B1/027-a brief overview[J]. Cureus, 2019, 11(1): e3977. |
| [58] | MARTÍNEZ-MELÉNDEZ A, CRUZ-LÓPEZ F, MORFIN-OTERO R, MALDONADO-GARZA HJ, GARZA-GONZÁLEZ E. An update on Clostridioides difficile binary toxin[J]. Toxins, 2022, 14(5): 305 DOI:10.3390/toxins14050305. |
| [59] | BILVERSTONE TW, MINTON NP, KUEHNE SA. Phosphorylation and functionality of CdtR in Clostridium difficile[J]. Anaerobe, 2019, 58: 103-109 DOI:10.1016/j.anaerobe.2019.102074. |
| [60] | YOUNG MK, LESLIE JL, MADDEN GR, LYERLY DM, CARMAN RJ, LYERLY MW, STEWART DB, ABHYANKAR MM, PETRI WA. Binary toxin expression by Clostridioides difficile is associated with worse disease[J]. Open Forum Infectious Diseases, 2022, 9(3): ofac001 DOI:10.1093/ofid/ofac001. |
| [61] | COSTA DVS, PHAM NVS, HAYS RA, BOLICK DT, GOLDBECK SM, POULTER MD, HOANG SC, SHIN JH, WU M, WARREN CA. Influence of binary toxin gene detection and decreased susceptibility to antibiotics among Clostridioides difficile strains on disease severity: a single-center study[J]. Antimicrobial Agents and Chemotherapy, 2022, 66(8): e0048922 DOI:10.1128/aac.00489-22. |
| [62] | PAPARELLA AS, ABOULACHE BL, HARIJAN RK, POTTS KS, TYLER PC, SCHRAMM VL. Inhibition of Clostridium difficile TcdA and TcdB toxins with transition state analogues[J]. Nature Communications, 2021, 12: 6285 DOI:10.1038/s41467-021-26580-6. |
 2023, Vol. 63
2023, Vol. 63