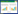中国科学院微生物研究所,中国微生物学会
文章信息
- 杨裕然, 李辉妙, 李振轮, 汪恩旭. 2023
- YANG Yuran, LI Huimiao, LI Zhenlun, WANG Enxu.
- 好氧氨氧化过程中的关键酶及N2O排放研究进展
- Key enzymes and N2O emission in aerobic ammonia oxidation process
- 微生物学报, 63(9): 3321-3334
- Acta Microbiologica Sinica, 63(9): 3321-3334
-
文章历史
- 收稿日期:2022-12-22
- 网络出版日期:2023-02-15
2. 西南大学植物保护学院 植物病害生物学重庆市高校重点实验室, 重庆 400716
2. Chongqing Key Laboratory of Plant Disease Biology, College of Plant Protection, Southwest University, Chongqing 400716, China
一氧化二氮(nitrous oxide, N2O)是大气中的一种长寿命痕量温室气体,目前在大气中的寿命为(116±9)年[1]。N2O成为了继CO2和CH4之后第三大最重要的温室气体,对全球变暖的贡献高达6%[2]。同时,N2O的增温潜势分别约为CO2和CH4的300倍和10倍[2-3]。在过去的40年中,N2O排放量增加了30%以上[4],并且人为因素增加的排放量占全球总量的30%–45%[5]。因此,更好地研究微生物N2O产生过程对实现人工调控减少N2O排放、减缓温室效应、维护地球生态平衡均具有重要意义。
微生物N2O排放的主要过程包括硝化作用、硝化反硝化作用和反硝化作用[1, 6]。如图 1所示,硝化作用是微生物将铵(NH4+)氧化为硝酸盐(NO3–),N2O作为副产物排放出来。硝化反硝化是将亚硝酸盐(NO2–)还原为一氧化氮(NO),最后转化为N2O或N2。反硝化是将NO3–转化为N2O,最后转化为惰性N2。而硝化过程中的好氧氨氧化形成的NO2–或NO3–为异养反硝化过程提供底物,因此好氧氨氧化直接或间接地影响着全球产生N2O与释放量[7]。Hink等[8]证实好氧氨氧化是土壤N2O排放的主要原因。好氧氨氧化菌分为自养和异养两大类,自养氨氧化菌包括氨氧化古菌(ammonia-oxidizing archaea, AOA)、氨氧化细菌(ammonia-oxidizing bacteria, AOB)和新发现的全程氨氧化菌(complete ammonia oxidization, Comammox)。因早期认为自然界异养氨氧化弱,因此好氧氨氧化及形成N2O的途径和机理主要依据自养氨氧化菌的研究结果。
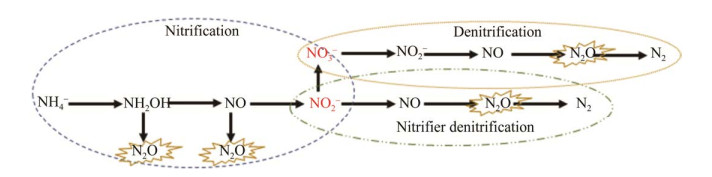
|
| 图 1 微生物N2O排放过程 Figure 1 Microbial N2O emission process. |
1 自养氨氧化菌的好氧氨氧化过程及N2O排放
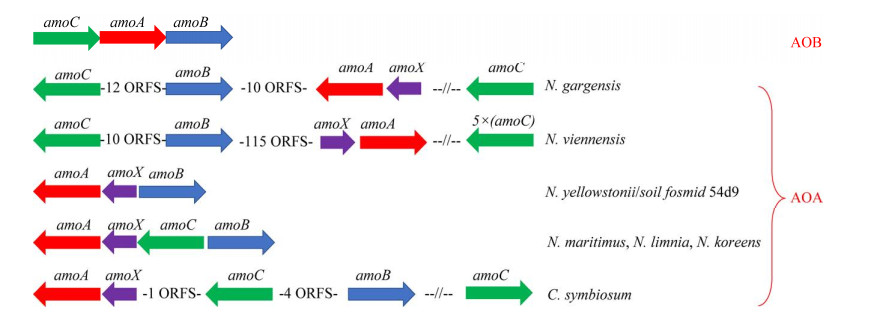
好氧氨氧化是硝化作用的一个限速步骤。在这个过程中AOA、AOB和Comammox都使用保守的氨单加氧酶(ammonia monooxygenase, AMO)将氨(NH3而不是NH4+)氧化成羟胺(NH2OH) [公式(1)];进一步地将NH2OH氧化成NO2–,但该途径所涉及的酶学和中间反应过程仍未完全解决,有待进一步研究。但总的来说AMO是三大类氨氧化微生物(AOA、AOB和Comammox)中氨氧化途径唯一共有的酶,NH2OH和NO是必经的中间产物[9-11]。
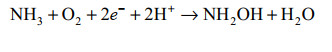
|
公式(1) |
好氧氨氧化的第一步是NH3在AMO的作用下氧化为NH2OH。通过16S rRNA和amoA基因发现,AMO是AOA[12]、AOB[13]和Comammox[14-16]这三类氨氧化微生物唯一所共有的关键酶。AMO是一种铜依赖性多聚体跨膜酶,与颗粒甲烷单加氧酶(particulate methane monooxygenase, pMMO)属于同一铜依赖性膜单加氧酶超家族[11]。自养氨氧化菌的AMO在蛋白提取液中稳定性差,尽管其细胞裂解物的体外活性可以在某些条件维持(如避光、添加Mg2+和牛血清蛋白等)[17-18],但AMO全酶尚未以纯化的活性形式分离出来,这极大地阻碍了对该蛋白质的结构和机制知识的扩展[19-20]。关于AMO结构和功能的见解主要是从全细胞或细胞提取液的实验中推导而来[21]或基于与其同源性较好的pMMO推导而来的[22]。例如,细菌好氧氨氧化成羟胺的活性被铜螯合剂烯丙基硫脲(ATU)抑制;少量Cu的加入可以明显刺激氨氧化,并能稳定细胞提取液中AMO的活性,这表明AMO是一种铜依赖性酶[23]。然而硝化抑制剂对AOA和AOB的活性影响存在差异,这表明AOA和AOB在AMO结构和功能上存在差异[24]。
AMO为多聚体膜结合蛋白,目前已分离到的确定为AMO亚基的有3个(amoA、amoB和amoC),并且在所有的AOB都以amoC、amoA、amoB的顺序排列在同一操纵子中[10, 22, 25]。此外,在amoCAB操纵子的下游也有关于AmoD和AmoE的报道[26]。相关研究认为,amoA和amoC亚基是完整的膜蛋白,而amoB亚基包含铜结合催化位点[27]。由于amoD和amoE的确切功能未知,因此amoCABED基因簇通常被称为amoCAB操纵子。amoA基因是最保守的亚基,常被用于其他亚群中系统发育研究[21, 28-29]。当然也存在其他操纵子的可能性。如,El Sheikh等[30]研究发现,在氨氧化细菌Nitrosococcus oceani ATCC 19707的amoCAB操纵子的上游和下游分别存在另外的2个保守基因amoR和amoD,形成了特异性amoRCABD操纵子。Bollmann等[26]报道了在Nitrosomonas sp. Is79的amoCAB操纵子的下游存在2个单拷贝的amoC基因。Kozlowski等[31]通过对比5株AOB发现都含有1–2个编码AMO的amoCABED簇。
在AOA的AMO中除了常见的amoA、amoB和amoC亚基外,通常还包括amoX[28, 32],并且这4个亚基的顺序在不同的AOA中的排列顺序多变。一些菌的亚基基因按amoAXCB排列(图 2)。一些AOA包含多个分离的amoA、amoB和amoC亚基副本。同时AOA中amoC的氮端和amoA的碳端比AOB短[22]。也正是因为AOA的AMO亚基排列顺序与AOB的AMO亚基排列顺序的不同,有人认为AOA氨氧化的产物可能不是NH2OH而是亚硝酸(nitroxyl, HNO),但缺乏直接的证据[28, 33]。
迄今为止发现的所有可培养的Comammox都是自养型微生物,属于Nitrospira[14, 34],从完全氨氧化为硝酸盐中获得能量[29]。与AOA和AOB的amoA基因相比,Comammox的amoA基因具有不同的基因序列[35-36],表现出更高的多样性[37]。但与颗粒甲烷单加氧酶基因pmoA更为相同[16]。Comammox中的AMO由单个amoCAB基因簇和其他基因组位点的2个额外amoC基因编码[14]。由于目前分离纯化得到的Comammox菌株较少,多通过组装基因组学获得相关基因,并且多关注于amoA,因此有关Comammox的AMO的其他性质有待进一步研究。
1.2 羟胺氧化为NO2–在不同的好氧氨氧化菌中存在差异,但NH2OH和NO是必经中间产物 1.2.1 AOB中羟胺经HAO转化为NO
好氧氨氧化的普遍观点主要基于对AOB亚硝化单胞菌模型的研究,通常认为氨氧化的第二步是NH2OH在羟胺氧化还原酶(hydroxylamine oxidoreductase, HAO)的作用下转化为NO2–,并产生4个电子,其中2个返回到AMO [公式(2)][38-39]。随着Caranto和Lancaster[40]研究结果的发现打破了这一定论(图 3)。他们认为羟胺受到HAO和细胞素色P460(厌氧)的共同作用。当NH2OH受到HAO和细胞素色P460的催化时分别产生3个和2个电子,并且形成的直接产物是NO而不是NO2–[40]。羟胺在HAO的催化下产生NO和3个电子,其中的2个电子回到AMO,1个电子进入细胞呼吸[公式(3)]。接着NO在一氧化氮氧化还原酶(nitric oxide oxidoreductase, NOO)的作用下氧化为NO2–[公式(4)]。最近通过对比不同AOB研究发现,亚硝基蓝蛋白(nitrosocyanin, NcyA)在氨氧化过程中同AMO和HAO一样高度表达,并能够与NO结合,被认为是缺失的第3种酶[13, 41-42]。发现NcyA参与将电子从醌池循环到AMO或作为羟胺到氧气的电子中继器[13]。但是在Nitrosomonas sp. Is 79的全基因组中没有发现ncyA基因[26]。此外,也有人认为是由反向操作的含铜亚硝酸盐还原酶(copper nitrite reductase, Cu-NirK)行使由NO到NO2–的功能[42]。但在某些AOB的全基因组中也找不到相关基因[41-42]。因此有必要进一步研究在AOB中行使功能的第3种酶。
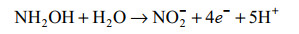
|
公式(2) |
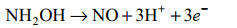
|
公式(3) |
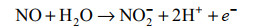
|
公式(4) |
在AOB的羟胺氧化过程中确定功能的蛋白只有HAO。由于HAO可溶性质,其是氨氧化过程中研究最清楚的功能组分。HAO蛋白已结晶,其结构也已解析。HAO是一种多血红素酶,由同质三聚亚基组成,每个亚基的分子量为67.1 kDa[43]。编码hao的基因表达为单顺反子转录本。从N. eurpaea中鉴定出3个hao基因拷贝:hao1、hao2和hao3。并且hao1和hao2的核酸序列几乎相同,而hao3与其他两个存在差异[44]。HAO酶的每个亚基包含8个c型血红素:7个是电子转移辅助因子,第8个是由细胞色素P460组成的羟胺氧化的活性位点[44]。来自HAO的电子流通过细胞色素c554被传至细胞色素cm552[45]。值得注意的,最近的一项新研究发现,DnfA编码的羟胺氧化酶在有氧的条件下能将羟胺直接转化为N2[46]。
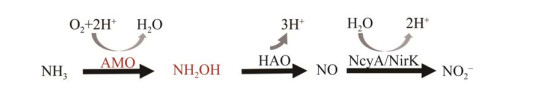
1.2.2 AOA中羟胺经含铜的未知酶转化为NO在AOA好氧氨氧化过程中也观测到了NO的产生[47-48],表明NO是该过程中的中间产物。通过添加NO的清除剂(如PTIO)显著抑制AOA氨氧化形成NO2–[41]。但由于AOA基因组中缺乏任何可识别的与AOB中HAO相近的同源物,这意味着AOA中有其他酶来负责羟胺氧化[5, 22]。并且进一步由NO氧化为NO2–所涉及的酶也有待进一步确定[39, 41]。目前,提出了2个可能的AOA羟胺氧化模型。一种模型认为羟胺和NO共同作用催化产生2分子NO2– (图 4A)[47]。另一种模型是羟胺连续通过2种酶先氧化为NO再氧化为NO2– (图 4B)[49]。羟胺通过未知酶氧化产生5个电子。其中2个电子被运送到膜相关醌还原酶(membrane-associated quinone reductase, QUED),然后通过一个蓝色的含铜蛋白载体的作用转移到AOA的AMO进行氨氧化[50-51]。剩余的电子被转移到电子传递链上用于呼吸。目前认为NH2OH氧化为NO2–过程中涉及的酶必须是铜基的[41]。有两位可能的候选者,即亚硝酸盐铜还原酶(copper nitrite reductase, Cu-NirK)和铜羟胺氧化还原酶(copper hydroxylamine oxidoreductase, Cu-HAO)[28]。由于Cu-HAO与AOB的HAO是同源物,但编码Cu-HAO的基因尚未被识别[28, 33],因此不确定AOA中HAO是否存在。而Kobayashi等[52]通过异源表达NirK发现,重组NirK蛋白能够催化NO2–还原为NO和催化羟胺氧化为NO。因此,Cu-NirK认为可能是与细菌HAO的对应物。但现有推导的古菌NirK与细菌NirK的相似度较低,确切的生理作用尚不确定[28],因此这也是今后需要解决的问题。
1.2.3 Comammox中羟胺经未知酶转化为NO
Comammox菌在好氧氨氧化过程中产NO也得到了验证[5]。目前认为Comammox的羟胺氧化过程与AOB类似。Dimitri Kits等[5]通过对宏基因组组装基因组分析发现,15个Comammox菌均含有HAO酶,并且推断羟胺在HAO酶的作用下产生NO。并且在一种未知的NOO酶作用下将NO氧化为NO2–。但是并未在Comammox菌的基因组中找到类似AOB中的ncyA基因[5]。因此,有其他的基因行使NOO酶的功能。此外,Dimitri Kits等[5]在所有Comammox菌的基因组中发现了nirK基因,但是认为在细胞内pH值和氧化还原电位下,反应动力学非常不利,表明NirK不是NOO的理想候选物。总之,Comammox菌有通过HAO或NirK产生NO的遗传潜力,但缺乏形成N2O的关键酶。
1.3 自养好氧氨氧化过程中N2O的排放大量研究证实AOA和AOB是N2O的主要产生源。而Comammox介导的过程被认为是“绿色过程”[36, 53],其对N2O排放量的贡献没有明确的报告,即使有产生N2O,量也特别少[54]。因此,主要讨论AOA和AOB在好氧氨氧化过程中涉及的N2O排放。
好氧氨氧化过程中参与N2O排放的物质有NH2OH、NO和NO2–。(1) NH2OH以及NH2OH和NO共同在细胞色素P460的作用下直接转化为N2O是在厌氧情况下发生的[55-56],不在本次的讨论范围。(2) NO经一氧化氮还原酶(nitric oxide reductase, NOR)产生N2O似乎是AOB所独有的过程[57]。目前在所有的AOA[47]和Comammox菌[16]中都没有发现nor基因的同源物。(3) NO2–经过硝化反硝化过程逐步产生NO和N2O。由于AOA缺乏NOR酶,通过同位素研究发现N2O中的N来自NH4+和NO2–[33]。因此认为AOA中的N2O是由非生物反应产生的[31, 47]。此外,Comammox菌是较新发现的,有关其N2O排放的研究较少。Dimitri Kits等[5]发现Comammox菌对NO清除剂很敏感,不能反硝化为N2O,并且不含有nor基因。因此Comammox菌在好氧氨氧化过程中的N2O排放是非生物转化的[36]。总的来说,好氧氨氧化过程中生物N2O排放关键酶是NOR,其催化NO到N2O的转化。
2 异养氨氧化菌的好氧氨氧化过程及N2O异养氨氧化早在一百多年前就有报道。近年来,异养氨氧化菌(heterotrophic ammonium oxidizing bacteria, HAOB)因其在酸性土壤氨氧化及其产生N2O量中起重要作用而得到极大关注[58]。已从旱地、水田、废水处理厂和养殖场等多种场所分离纯化种类繁多的异养硝化菌,对氮转化途径及特性、影响因素和功能应用等方面进行了详细研究[59-63]。
2.1 异养氨氧化菌的好氧氨氧化过程存在差异相较于自养氨氧化菌,HAOB的底物范围广泛[56]。并且异养氨氧化往往与好氧反硝化耦联发生[64],这使得从中间产物推断异养硝化机理异常困难。目前对HAOB好氧氨氧化途径一般是基于不同氮源的利用和代谢产物、功能酶或基因的检测而提出的[63]。但异养氨氧化发生的代谢途径没有共识。即使是同一种属的HAOB好氧氨氧化过程都存在差异。例如,有研究认为假单胞菌(Pseudomonas sp.)的好氧氨氧化途径为NH4+→NH2OH→NO2–→NO3–→ NO2–→NO→N2O→N2[65-66],也有研究认为Pseudomonas sp.的好氧氨氧化途径为NH4+→NH2OH→NO2–→气态氮[67]。但是,也有学者认为异养硝化跨过了NH4+分解,即有机氮→NH2OH→NO2–→NO→N2O→N2[68]。值得注意的是,通过基因推测好氧氨氧化过程似乎也存在问题。Silva等[69]对具有氨氧化能力的菌株Pseudomonas stutzeri进行测序发现,基因中未检测到与自养硝化菌氨氧化过程中已知的基因和酶,但发现反硝化过程中所需的基因和酶存在。
2.2 异养氨氧化菌好氧氨氧化过程的酶学不完全清楚HAOB好氧氨氧化过程的基因和酶学是不完善的,那么对HAOB好氧氨氧化第一步是否由AMO催化产生NH2OH存在质疑。一方面由于NH2OH含量低较难检测到,如在Streptomyces mediolani EM-B2[23]、P. aeruginosa[64]、Pseudomonas putida Y-9[70]和Acinetobacter sp. ND7[71]氨氧化过程中均没有检测到NH2OH的存在。那么推断可能的氨氧化过程为NH4+→NO2–→NO→N2O→ N2[72]。另一方面异养氨氧化过程是基于自养氨氧化过程推测的,普遍接受了氨在AMO的作用下产生NH2OH。但目前仅在少数HAOB中发现了AMO。如Lang等[73]使用amoA基因在3株HAOB中扩增出AMO条带。虽然Wang等[74]也扩增出AMO条带,但amoA引物来自自身高通量数据。相较于自养氨氧化菌,HAOB的AMO在细胞提取液中稳定,由2个亚基[75],或单一蛋白[18]组成。但值得注意的是,也有一些异养硝化菌不含amoA基因或基因组缺乏传统硝化途径关键基因[76]。例如,本实验室前期在粗酶液中检测到AMO酶活,但使用AOB的amoA引物没有扩增出Pseudomonas putida Y-9的AMO条带[70]。Cui等[77]也是相同的结果。此外,现有的硝化抑制剂(主要针对自养氨氧化菌设计)对HAOB作用效果不明显[70, 78]。如本实验室发现,0.5 mmol/L ATU对AOB有显著的抑制作用,而对Arthrobacter arilaitensis无抑制作用;但随着浓度的增加,抑制作用增强[79]。而Pseudomonas putida Y-9对ATU极不敏感[70]。以上结果说明在某些异养氨氧化菌中可能存在不同于自养氨氧化菌的氨氧化过程。
总的来说,HAOB氨氧化过程所涉及的基因和酶都不是很清楚,可能针对不同种属的HAOB需要提出特定的氨氧化过程。
2.3 异养氨氧化过程中N2O的排放目前关于不同生态系统及体系中N2O的产生量及机理有很多报道,发现不少系统中异养硝化作用排放N2O强烈。如Liu等[80]研究发现异养硝化作用对水稻土释放N2O的贡献比自养硝化作用更大。Zhang等[81]使用15N同位素标记证明异养硝化作用会诱导我国亚热带酸性森林土壤和温带森林土壤产N2O,并且是亚热带酸性森林土壤产N2O的主要原因。Pan等[60]通过高通量测序发现异养硝化菌在处理高盐废水脱氮中发挥了关键作用。Fan等[82]研究表明,3, 4-二甲基吡唑磷酸酯(3, 4-dimethylpyrazole phosphate, DMPP,一种硝化抑制剂)对红壤产N2O仅能抑制41.7%,而对灰漠土抑制率高达90.0%。究其原因与酸性红壤含异养硝化菌多有关。异养硝化菌的氨氧化酶与自养菌不同,其活性对传统意义的硝化抑制剂可能不敏感[82-83]。
目前认为HAOB氨氧化排放N2O的途径有2条。(1) 与自养氨氧化类似,往往针对异养硝化和好氧反硝化相耦合的N2O排放:NH4+→NH2OH→NO2–→NO→N2O。如本实验室发现菌株Pseudomonas putida Y-9[70]在氨氧化过程中会有NO2–的积累,并且生产的气态氮均为N2O。(2) 是HAOB仅进行异养氨氧化的N2O排放途径:NH4+→NH2OH→NO→N2O。如Acinetobacter calcoaceticus HNR[84]能够去除NH4+和NH2OH,但不能利用NO2–和NO3–,故推测第(2)途径为可能的N2O排放途径。类似的还有Zhao等[59]报道的Alcaligenes faecalis NR,不从NO2–和NO3–产生N2O,而是NH2OH直接产生N2O。然而,异养硝化菌的氮代谢过程的关键基因及酶都是根据自养硝化菌的研究结果来推测的,异养氨氧化菌的AMO亚基组成、羟胺还原酶等的结构及生理作用都不清楚,无法从异养氨氧化产N2O机理方面进行分析,导致研究结果不能更加全面地反应氮代谢途经及其影响因素对N2O产生的贡献,对人们寻找减少N2O排放的有效方法带来极大的阻碍。
3 展望好氧氨氧化是氮循环过程中的一个重要过程,该过程直接或间接的影响着全球N2O的排放量。本文系统的综述了好氧氨氧化过程中参与的微生物,包括该过程中涉及的中间体、关键酶及N2O的排放问题。并且发现了现有研究中存在的不足,需要各位研究者进一步的探究:
(1) 羟胺氧化的直接产物NO已经得到证实,但有关从NO到NO2–的过程中所涉及的酶及基因还有待进一步的确定。
(2) 新发现的Comammox菌的大部分基因没有功能注释,缺乏对相关蛋白的纯化和表征,需要进行生理实验验证;并且16S rRNA测序无法区分Comammox和亚硝酸盐还原菌[39, 85],需要针对Comammox设计专门的引物。
(3) 异养氨氧化菌可能具有氨氧化酶,但现有AMO引物不能很好的对其进行表征[63]。因此需要针对异养氨氧化菌开发特异引物;此外,异养氨氧化菌种类繁多,过程复杂,往往与其他过程同时发生(如好氧反硝化、碳循环等),并且相关酶学和基因不清楚,而其在全球N2O排放的贡献不可忽略。而现有的硝化抑制剂对其效果不显著。因此需要研发针对异养氨氧化菌的特异性抑制剂。
| [1] | TIAN HQ, XU RT, CANADELL JG, THOMPSON RL, WINIWARTER W, SUNTHARALINGAM P, DAVIDSON EA, CIAIS P, JACKSON RB, JANSSENS-MAENHOUT G, PRATHER MJ, REGNIER P, PAN NQ, PAN SF, PETERS GP, SHI H, TUBIELLO FN, ZAEHLE S, ZHOU F, ARNETH A, et al. A comprehensive quantification of global nitrous oxide sources and sinks[J]. Nature, 2020, 586(7828): 248-256 DOI:10.1038/s41586-020-2780-0. |
| [2] | SHAKOOR A, SHAKOOR S, REHMAN A, ASHRAF F, ABDULLAH M, SHAHZAD SM, FAROOQ TH, ASHRAF M, MANZOOR MA, ALTAF MM, ALTAF MA. Effect of animal manure, crop type, climate zone, and soil attributes on greenhouse gas emissions from agricultural soils—a global meta-analysis[J]. Journal of Cleaner Production, 2021, 278: 124019 DOI:10.1016/j.jclepro.2020.124019. |
| [3] | SOLER-JOFRA A, PICIOREANU C, YU R, CHANDRAN K, van LOOSDRECHT MCM, PÉREZ J. Importance of hydroxylamine in abiotic N2O production during transient anoxia in planktonic axenic Nitrosomonas cultures[J]. Chemical Engineering Journal, 2018, 335: 756-762 DOI:10.1016/j.cej.2017.10.141. |
| [4] | ZHANG Y, ZHANG F, ABALOS D, LUOYQ, HUI DF, HUNGATE BA, GARCÍA-PALACIOS P, KUZYAKOV Y, OLESEN JE, JØRGENSEN U, CHEN J. Stimulation of ammonia oxidizer and denitrifier abundances by nitrogen loading: poor predictability for increased soil N 2 O emission[J]. Global Change Biology, 2022, 28(6): 2158-2168 DOI:10.1111/gcb.16042. |
| [5] | DIMITRI KITS K, JUNG MY, VIERHEILIG J, PJEVAC P, SEDLACEK CJ, LIU SR, HERBOLD C, STEIN LY, RICHTER A, WISSEL H, BRÜGGEMANN N, WAGNER M, DAIMS H. Low yield and abiotic origin of N2O formed by the complete nitrifier Nitrospira inopinata[J]. Nature Communications, 2019, 10: 1836 DOI:10.1038/s41467-019-09790-x. |
| [6] | WANG C, AMON B, SCHULZ K, MEHDI B. Factorsthat influence nitrous oxide emissions from agricultural soils as well as their representation in simulation models: a review[J]. Agronomy, 2021, 11(4): 770 DOI:10.3390/agronomy11040770. |
| [7] | ELOY ALVES RJ, MINH BQ, URICH T, von HAESELER A, SCHLEPER C. Unifying the global phylogeny and environmental distribution of ammonia-oxidising archaea based on amoA genes[J]. Nature Communications, 2018, 9: 1517 DOI:10.1038/s41467-018-03861-1. |
| [8] | HINK L, NICOL GW, PROSSER JI. Archaea produce lower yields of N2O than bacteria during aerobic ammonia oxidation in soil[J]. Environmental Microbiology, 2017, 19(12): 4829-4837 DOI:10.1111/1462-2920.13282. |
| [9] | BEECKMAN F, MOTTE H, BEECKMAN T. Nitrification in agricultural soils: impact, actors and mitigation[J]. Current Opinion in Biotechnology, 2018, 50: 166-173 DOI:10.1016/j.copbio.2018.01.014. |
| [10] | CHAWLEY P, RANAAN, JAGADEVAN S. Envisioning role of ammonia oxidizing bacteria in bioenergy production and its challenges: a review[J]. Critical Reviews in Biotechnology, 2022, 42(6): 931-952 DOI:10.1080/07388551.2021.1976099. |
| [11] | WRIGHT CL, SCHATTEMAN A, CROMBIE AT, MURRELL JC, LEHTOVIRTA-MORLEY LE. Inhibition of ammonia monooxygenase from ammonia-oxidizing archaea by linear and aromatic alkynes[J]. Applied and Environmental Microbiology, 2020, 86(9): e02388-e02319. |
| [12] | PESTER M, RATTEI T, FLECHL S, GRÖNGRÖFT A, RICHTER A, OVERMANN J, REINHOLD-HUREK B, LOY A, WAGNER M. amoA-based consensus phylogeny of ammonia-oxidizing archaea and deep sequencing of amoA genes from soils of four different geographic regions[J]. Environmental Microbiology, 2012, 14(2): 525-539 DOI:10.1111/j.1462-2920.2011.02666.x. |
| [13] | ZORZ JK, KOZLOWSKI JA, STEIN LY, STROUS M, KLEINER M. Comparativeproteomics of three species of ammonia-oxidizing bacteria[J]. Frontiers in Microbiology, 2018, 9: 938 DOI:10.3389/fmicb.2018.00938. |
| [14] | DAIMS H, LEBEDEVA EV, PJEVAC P, HAN P, HERBOLD C, ALBERTSEN M, JEHMLICH N, PALATINSZKY M, VIERHEILIG J, BULAEV A, KIRKEGAARD RH, von BERGEN M, RATTEI T, BENDINGER B, NIELSEN PH, WAGNER M. Complete nitrification by Nitrospira bacteria[J]. Nature, 2015, 528(7583): 504-509 DOI:10.1038/nature16461. |
| [15] | van KESSEL MAHJ, SPETH DR, ALBERTSEN M, NIELSEN PH, den CAMP HJMO, KARTAL B, JETTEN MSM, LÜCKER S. Complete nitrification by a single microorganism[J]. Nature, 2015, 528(7583): 555-559 DOI:10.1038/nature16459. |
| [16] | PALOMO A, PEDERSEN AG, FOWLER SJ, DECHESNE A, SICHERITZ-PONTÉN T, SMETS BF. Comparative genomics sheds light on niche differentiation and the evolutionary history of comammox Nitrospira[J]. The ISME Journal, 2018, 12(7): 1779-1793 DOI:10.1038/s41396-018-0083-3. |
| [17] | ENSIGN SA, HYMAN MR, ARP DJ. In vitro activation of ammonia monooxygenase from Nitrosomonas europaea by copper[J]. Journal of Bacteriology, 1993, 175(7): 1971-1980 DOI:10.1128/jb.175.7.1971-1980.1993. |
| [18] | ZHANGSM, LI WG, ZHANG DY, HUANG XF, QIN W, GU J. Purification and characterization of a low-temperature ammonia monooxygenase from heterotrophic nitrifier Acinetobacter sp. Y16[J]. Desalination and Water Treatment, 2015, 53(1): 257-262 DOI:10.1080/19443994.2013.837002. |
| [19] | MUSIANI F, BROLL V, EVANGELISTI E, CIURLI S. The model structure of the copper-dependent ammonia monooxygenase[J]. JBIC Journal of Biological Inorganic Chemistry, 2020, 25(7): 995-1007 DOI:10.1007/s00775-020-01820-0. |
| [20] | FISHER OS, KENNEY GE, ROSS MO, RO SY, LEMMA BE, BATELU S, THOMAS PM, SOSNOWSKI VC, DeHART CJ, KELLEHER NL, STEMMLER TL, HOFFMAN BM, ROSENZWEIG AC. Characterization of a long overlooked copper protein from methane- and ammonia-oxidizing bacteria[J]. Nature Communications, 2018, 9: 4276 DOI:10.1038/s41467-018-06681-5. |
| [21] | GUO JH, PENG YZ, WANG SY, MA B, GE SJ, WANG ZW, HUANG HJ, ZHANG JR, ZHANG L. Pathways and organisms involved in ammonia oxidation and nitrous oxide emission[J]. Critical Reviews in Environmental Science and Technology, 2013, 43(21): 2213-2296 DOI:10.1080/10643389.2012.672072. |
| [22] | LEHTOVIRTA-MORLEY LE. Ammonia oxidation: ecology, physiology, biochemistry and why they must all come together[J]. FEMS Microbiology Letters, 2018, 365(9): fny058. |
| [23] | HETX, ZHANG MM, DING CY, WU QF, CHEN MP, MOU SL, CHENG DJ, DUAN SJ, WANG Y. New insight into the nitrogen removal capacity and mechanism of Streptomyces mediolani EM-B2[J]. Bioresource Technology, 2022, 348: 126819 DOI:10.1016/j.biortech.2022.126819. |
| [24] | SHENTL, STIEGLMEIER M, DAI JL, URICH T, SCHLEPER C. Responses of the terrestrial ammonia-oxidizing archaeon Ca. Nitrososphaera viennensis and the ammonia-oxidizing bacterium Nitrosospira multiformis to nitrification inhibitors[J]. FEMS Microbiology Letters, 2013, 344(2): 121-129 DOI:10.1111/1574-6968.12164. |
| [25] | TAVORMINA PL, ORPHAN VJ, KALYUZHNAYA MG, JETTENMSM, KLOTZ MG. A novel family of functional operons encoding methane/ammonia monooxygenase-related proteins in gammaproteobacterial methanotrophs[J]. Environmental Microbiology Reports, 2011, 3(1): 91-100 DOI:10.1111/j.1758-2229.2010.00192.x. |
| [26] | BOLLMANN A, SEDLACEK CJ, NORTON J, LAANBROEK HJ, SUWA Y, STEIN LY, KLOTZ MG, ARP D, SAYAVEDRA-SOTO L, LUMG, BRUCE D, DETTER C, TAPIA R, HAN J, WOYKE T, LUCAS SM, PITLUCK S, PENNACCHIO L, NOLAN M, LAND ML, et al. Complete genome sequence of Nitrosomonas sp. Is79, an ammonia oxidizing bacterium adapted to low ammonium concentrations[J]. Standards in Genomic Sciences, 2013, 7(3): 469-482. |
| [27] | BALASUBRAMANIAN R, SMITH SM, RAWAT S, YATSUNYK LA, STEMMLER TL, ROSENZWEIG AC. Oxidation of methane by a biological dicopper centre[J]. Nature, 2010, 465(7294): 115-119 DOI:10.1038/nature08992. |
| [28] | WU L, CHENXM, WEI W, LIU YW, WANG DB, NI BJ. A critical review on nitrous oxide production by ammonia-oxidizing archaea[J]. Environmental Science & Technology, 2020, 54(15): 9175-9190. |
| [29] | AL-AJEEL S, SPASOV E, SAUDER LA, McKNIGHT MM, NEUFELD JD. Ammonia-oxidizing archaea and complete ammonia-oxidizing Nitrospira in water treatment systems[J]. Water Research X, 2022, 15: 100131 DOI:10.1016/j.wroa.2022.100131. |
| [30] | EL SHEIKH AF, PORET-PETERSON AT, KLOTZ MG. Characterization of two new genes, amoR and amoD, in the Amo operon of the marine ammonia oxidizer Nitrosococcus oceani ATCC 19707[J]. Applied and Environmental Microbiology, 2008, 74(1): 312-318 DOI:10.1128/AEM.01654-07. |
| [31] | KOZLOWSKI JA, KITS KD, STEIN LY. Comparison of nitrogen oxide metabolism among diverse ammonia-oxidizing bacteria[J]. Frontiers in Microbiology, 2016, 7: 1090. |
| [32] | KEROU M, OFFRE P, VALLEDOR L, ABBY SS, MELCHER M, NAGLER M, WECKWERTH W, SCHLEPER C. Proteomics and comparative genomics of Nitrososphaera viennensis reveal the core genome and adaptations of archaeal ammonia oxidizers[J]. Proceedings of the National Academy of Sciences of the United States of America, 2016, 113(49): E7937-E7946. |
| [33] | STIEGLMEIER M, MOOSHAMMER M, KITZLER B, WANEK W, ZECHMEISTER-BOLTENSTERN S, RICHTER A, SCHLEPER C. Aerobic nitrous oxide production through N-nitrosating hybrid formation in ammonia-oxidizing archaea[J]. The ISME Journal, 2014, 8(5): 1135-1146 DOI:10.1038/ismej.2013.220. |
| [34] | SPASOV E, TSUJI JM, HUG LA, DOXEY AC, SAUDER LA, PARKER WJ, NEUFELD JD. High functional diversity among Nitrospira populations that dominate rotating biological contactor microbial communities in a municipal wastewater treatment plant[J]. The ISME Journal, 2020, 14(7): 1857-1872 DOI:10.1038/s41396-020-0650-2. |
| [35] | PJEVAC P, SCHAUBERGER C, POGHOSYAN L, HERBOLD CW, van KESSEL MAHJ, DAEBELER A, STEINBERGER M, JETTEN MSM, LÜCKER S, WAGNER M, DAIMS H. AmoA-targeted polymerase chain reaction primers for the specific detection and quantification of comammox Nitrospira in the environment[J]. Frontiers in Microbiology, 2017, 8: 1508 DOI:10.3389/fmicb.2017.01508. |
| [36] | ZHUGB, WANG XM, WANG SY, YU LB, ARMANBEK G, YU J, JIANG LP, YUAN DD, GUO ZR, ZHANG HR, ZHENG L, SCHWARK L, JETTEN MSM, YADAV AK, ZHU YG. Towards a more labor-saving way in microbial ammonium oxidation: a review on complete ammonia oxidization (comammox)[J]. Science of the Total Environment, 2022, 829: 154590 DOI:10.1016/j.scitotenv.2022.154590. |
| [37] | KOCH H, van KESSEL MAHJ, LÜCKER S. Complete nitrification: insights into the ecophysiology of comammox Nitrospira[J]. Applied Microbiology and Biotechnology, 2019, 103(1): 177-189 DOI:10.1007/s00253-018-9486-3. |
| [38] | CARANTO JD, LANCASTER KM. Correction for Caranto and Lancaster, nitric oxide is an obligate bacterial nitrification intermediate produced by hydroxylamine oxidoreductase[J]. Proceedings of the National Academy of Sciences of the United States of America, 2018, 115(35): E8325. |
| [39] | SOLER-JOFRA A, PÉREZ J, van LOOSDRECHT MCM. Hydroxylamine and the nitrogen cycle: a review[J]. Water Research, 2021, 190: 116723 DOI:10.1016/j.watres.2020.116723. |
| [40] | CARANTO JD, LANCASTER KM. Nitric oxide is an obligate bacterial nitrification intermediate produced by hydroxylamine oxidoreductase[J]. Proceedings of the National Academy of Sciences of the United States of America, 2017, 114(31): 8217-8222. |
| [41] | STEIN LY. Insights into the physiology of ammonia-oxidizing microorganisms[J]. Current Opinion in Chemical Biology, 2019, 49: 9-15 DOI:10.1016/j.cbpa.2018.09.003. |
| [42] | LANCASTER KM, CARANTO JD, MAJER SH, SMITH MA. Alternativebioenergy: updates to and challenges in nitrification metalloenzymology[J]. Joule, 2018, 2(3): 421-441 DOI:10.1016/j.joule.2018.01.018. |
| [43] | CEDERVALL P, HOOPER AB, WILMOT CM. Structuralstudies of hydroxylamine oxidoreductase reveal a unique heme cofactor and a previously unidentified interaction partner[J]. Biochemistry, 2013, 52(36): 6211-6218 DOI:10.1021/bi400960w. |
| [44] | ARP DJ, SAYAVEDRA-SOTO LA, HOMMES NG. Molecular biology and biochemistry of ammonia oxidation by Nitrosomonas europaea[J]. Archives of Microbiology, 2002, 178(4): 250-255 DOI:10.1007/s00203-002-0452-0. |
| [45] | UPADHYAY AK, HOOPER AB, HENDRICH MP. NO reductase activity of the tetraheme cytochrome C554 of Nitrosomonas europaea[J]. Journal of the American Chemical Society, 2006, 128(13): 4330-4337 DOI:10.1021/ja055183+. |
| [46] | WU MR, MIAO LL, LIU Y, QIAN XX, HOU TT, AI GM, YU L, MA L, GAO XY, QIN YL, ZHU HZ, DU L, LI SY, TIAN CL, LI DF, LIU ZP, LIU SJ. Identification and characterization of a novel hydroxylamine oxidase, DnfA, that catalyzes the oxidation of hydroxylamine toN2[J]. Journal of Biological Chemistry, 2022, 298(9): 102372 DOI:10.1016/j.jbc.2022.102372. |
| [47] | KOZLOWSKI JA, STIEGLMEIER M, SCHLEPER C, KLOTZ MG, STEIN LY. Pathways and key intermediates required for obligate aerobic ammonia-dependent chemolithotrophy in bacteria and Thaumarchaeota[J]. The ISME Journal, 2016, 10(8): 1836-1845 DOI:10.1038/ismej.2016.2. |
| [48] | MARTENS-HABBENA W, QIN W, HORAKREA, URAKAWA H, SCHAUER AJ, MOFFETT JW, ARMBRUST EV, INGALLS AE, DEVOL AH, STAHL DA. The production of nitric oxide by marine ammonia-oxidizing archaea and inhibition of archaeal ammonia oxidation by a nitric oxide scavenger[J]. Environmental Microbiology, 2015, 17(7): 2261-2274 DOI:10.1111/1462-2920.12677. |
| [49] | CARINI P, DUPONT CL, SANTORO AE. Patterns of thaumarchaeal gene expression in culture and diverse marine environments[J]. Environmental Microbiology, 2018, 20(6): 2112-2124 DOI:10.1111/1462-2920.14107. |
| [50] | WALKER CB, deLa TORRE JR, KLOTZ MG, URAKAWA H, PINEL N, ARP DJ, BROCHIER-ARMANET C, CHAIN PG, CHAN PP, GOLLABGIR A, HEMP J, HÜGLER M, KARR EA, KÖNNEKE M, SHIN M, LAWTON TJ, LOWE T, MARTENS-HABBENA W, SAYAVEDRA-SOTO LA, LANG D, et al. Nitrosopumilus maritimus genome reveals unique mechanisms for nitrification and autotrophy in globally distributed marine crenarchaea[J]. Proceedings of the National Academy of Sciences of the United States of America, 2010, 107(19): 8818-8823. |
| [51] | SPANG A, POEHLEIN A, OFFRE P, ZUMBRÄGEL S, HAIDER S, RYCHLIK N, NOWKA B, SCHMEISSER C, LEBEDEVA EV, RATTEI T, BÖHM C, SCHMID M, GALUSHKO A, HATZENPICHLER R, WEINMAIER T, DANIEL R, SCHLEPER C, SPIECK E, STREIT W, WAGNER M. The genome of the ammonia-oxidizing Candidatus Nitrososphaera gargensis: insights into metabolic versatility and environmental adaptations[J]. Environmental Microbiology, 2012, 14(12): 3122-3145 DOI:10.1111/j.1462-2920.2012.02893.x. |
| [52] | KOBAYASHI S, HIRA D, YOSHIDA K, TOYOFUKU M, SHIDA Y, OGASAWARA W, YAMAGUCHI T, ARAKI N, OSHIKI M. Nitricoxide production from nitrite reduction and hydroxylamine oxidation by copper-containing dissimilatory nitrite reductase (NirK) from the aerobic ammonia-oxidizing archaeon, Nitrososphaera viennensis[J]. Microbes and Environments, 2018, 33(4): 428-434 DOI:10.1264/jsme2.ME18058. |
| [53] | ANNAVAJHALA MK, KAPOOR V, SANTO-DOMINGO J, CHANDRAN K. Comammox functionality identified in diverse engineered biological wastewater treatment systems[J]. Environmental Science & Technology Letters, 2018, 5(2): 110-116. |
| [54] | DIMITRI KITS K, SEDLACEK CJ, LEBEDEVA EV, HAN P, BULAEV A, PJEVAC P, DAEBELER A, ROMANO S, ALBERTSEN M, STEIN LY, DAIMS H, WAGNER M. Kinetic analysis of a complete nitrifier reveals an oligotrophic lifestyle[J]. Nature, 2017, 549(7671): 269-272 DOI:10.1038/nature23679. |
| [55] | CARANTO JD, VILBERT AC, LANCASTER KM. Nitrosomonas europaea cytochrome P460 is a direct link between nitrification and nitrous oxide emission[J]. Proceedings of the National Academy of Sciences of the United States of America, 2016, 113(51): 14704-14709. |
| [56] | YU R, PEREZ-GARCIA O, LUHJ, CHANDRAN K. Nitrosomonas europaea adaptation to anoxic-oxic cycling: insights from transcription analysis, proteomics and metabolic network modeling[J]. Science of the Total Environment, 2018, 615: 1566-1573 DOI:10.1016/j.scitotenv.2017.09.142. |
| [57] | KOZLOWSKI JA, PRICE J, STEIN LY. Revision of N2O-producing pathways in the ammonia-oxidizing bacterium Nitrosomonas europaea ATCC 19718[J]. Applied and Environmental Microbiology, 2014, 80(16): 4930-4935 DOI:10.1128/AEM.01061-14. |
| [58] | CHENZM, DING WX, XU YH, MÜLLER C, RÜTTING T, YU HY, FAN JL, ZHANG JB, ZHU TB. Importance of heterotrophic nitrification and dissimilatory nitrate reduction to ammonium in a cropland soil: evidences from a 15N tracing study to literature synthesis[J]. Soil Biology and Biochemistry, 2015, 91: 65-75 DOI:10.1016/j.soilbio.2015.08.026. |
| [59] | ZHAO B, AN Q, HE YL, GUO JS. N2O and N2 production during heterotrophic nitrification by Alcaligenes faecalis strain NR[J]. Bioresource Technology, 2012, 116: 379-385 DOI:10.1016/j.biortech.2012.03.113. |
| [60] | PANZL, ZHOU J, LIN ZY, WANG YM, ZHAO PC, ZHOU J, LIU SH, HE XJ. Effects of COD/TN ratio on nitrogen removal efficiency, microbial community for high saline wastewater treatment based on heterotrophic nitrification-aerobic denitrification process[J]. Bioresource Technology, 2020, 301: 122726 DOI:10.1016/j.biortech.2019.122726. |
| [61] | YANG L, WANG XH, CUI S, REN YX, YU J, CHEN N, XIAO Q, GUO LK, WANG RH. Simultaneous removal of nitrogen and phosphorous by heterotrophic nitrification-aerobic denitrification of a metal resistant bacterium Pseudomonas putida strain NP5[J]. Bioresource Technology, 2019, 285: 121360 DOI:10.1016/j.biortech.2019.121360. |
| [62] | HEXL, SUN Q, XU TY, DAI M, WEI DS. Removal of nitrogen by heterotrophic nitrification-aerobic denitrification of a novel halotolerant bacterium Pseudomonas mendocina TJPU04[J]. Bioprocess and Biosystems Engineering, 2019, 42(5): 853-866 DOI:10.1007/s00449-019-02088-8. |
| [63] | DUANSP, ZHANG YY, ZHENG SK. Heterotrophic nitrifying bacteria in wastewater biological nitrogen removal systems: a review[J]. Critical Reviews in Environmental Science and Technology, 2022, 52(13): 2302-2338 DOI:10.1080/10643389.2021.1877976. |
| [64] | ZHU ZQ, YANG Y, FANG AR, LOU Y, XIE GJ, REN NQ, XING DF. Quorum sensing systems regulate heterotrophic nitrification-aerobic denitrification by changing the activity of nitrogen-cycling enzymes[J]. Environmental Science and Ecotechnology, 2020, 2: 100026 DOI:10.1016/j.ese.2020.100026. |
| [65] | ZHANG JB, LAN T, MÜLLER C, CAI ZC. Dissimilatory nitrate reduction to ammonium (DNRA) plays an important role in soil nitrogen conservation in neutral and alkaline but not acidic rice soil[J]. Journal of Soils and Sediments, 2015, 15(3): 523-531 DOI:10.1007/s11368-014-1037-7. |
| [66] | ZHOU MH, YE HR, ZHAO XW. Isolation and characterization of a novel heterotrophic nitrifying and aerobic denitrifying bacterium Pseudomonas stutzeri KTB for bioremediation of wastewater[J]. Biotechnology and Bioprocess Engineering, 2014, 19(2): 231-238 DOI:10.1007/s12257-013-0580-1. |
| [67] | LICE, YANG JS, WANG X, WANG ET, LI BZ, HE RX, YUAN HL. Removal of nitrogen by heterotrophic nitrification-aerobic denitrification of a phosphate accumulating bacterium Pseudomonas stutzeri YG-24[J]. Bioresource Technology, 2015, 182: 18-25 DOI:10.1016/j.biortech.2015.01.100. |
| [68] | ZHANG JB, MÜLLER C, CAI ZC. Heterotrophic nitrification of organic N and its contribution to nitrous oxide emissions in soils[J]. Soil Biology and Biochemistry, 2015, 84: 199-209 DOI:10.1016/j.soilbio.2015.02.028. |
| [69] | SILVA LCF, LIMA HS, de OLIVEIRA MENDES TA, SARTORATTO A, SOUSA MP, de SOUZA RS, de PAULA SO, de OLIVEIRA VM, SILVA CC. Physicochemical characterization of Pseudomonas stutzeri UFV5 and analysis of its transcriptome under heterotrophic nitrification/aerobic denitrification pathway induction condition[J]. Scientific Reports, 2020, 10: 2215 DOI:10.1038/s41598-020-59279-7. |
| [70] | HUANG XJ, WEISENER CG, NI JP, HE BH, XIE DT, LI ZL. Nitrate assimilation, dissimilatory nitrate reduction to ammonium, and denitrification coexist in Pseudomonas putida Y-9 under aerobic conditions[J]. Bioresource Technology, 2020, 312: 123597 DOI:10.1016/j.biortech.2020.123597. |
| [71] | XIA L, LIXM, FAN WH, WANG JL. Heterotrophic nitrification and aerobic denitrification by a novel Acinetobacter sp. ND7 isolated from municipal activated sludge[J]. Bioresource Technology, 2020, 301: 122749 DOI:10.1016/j.biortech.2020.122749. |
| [72] | LEI X, JIAYT, CHEN YC, HU YY. Simultaneous nitrification and denitrification without nitrite accumulation by a novel isolated Ochrobactrum anthropic LJ81[J]. Bioresource Technology, 2019, 272: 442-450 DOI:10.1016/j.biortech.2018.10.060. |
| [73] | LANG XD, LI QW, XU YC, JI MM, YAN GX, GUO SH. Aerobic denitrifiers with petroleum metabolizing ability isolated from caprolactam sewage treatment pool[J]. Bioresource Technology, 2019, 290: 121719 DOI:10.1016/j.biortech.2019.121719. |
| [74] | WANG XJ, WANG WQ, ZHANG Y, SUN ZT, ZHANG J, CHEN GH, LI J. Simultaneous nitrification and denitrification by a novel isolated Pseudomonas sp. JQ-H3 using polycaprolactone as carbon source[J]. Bioresource Technology, 2019, 288: 121506 DOI:10.1016/j.biortech.2019.121506. |
| [75] | MOIR JWB, CROSSMAN LC, SPIRO S, RICHARDSON DJ. The purification of ammonia monooxygenase from Paracoccus denitrficans[J]. FEBS Letters, 1996, 387(1): 71-74 DOI:10.1016/0014-5793(96)00463-2. |
| [76] | DOMINGUEZ MENDOZA LF, QUIMI MUJICA JG, RISCO CUNAYQUE JM, ARONI LUCANA GW, INTRIAGO ANGULO JJ, deLa CRUZ VIS, ALEXIA CEDEסO ESCOBAR V, MATONNIER EM. Assessment of heterotrophic nitrification capacity in Bacillus spp. and its potential application in the removal of nitrogen from aquaculture water[J]. Journal of Pure and Applied Microbiology, 2019, 13(4): 1893-1908 DOI:10.22207/JPAM.13.4.02. |
| [77] | CUI Y, CUI YW, HUANG JL. A novel halophilic Exiguobacterium mexicanum strain removes nitrogen from saline wastewater via heterotrophic nitrification and aerobic denitrification[J]. Bioresource Technology, 2021, 333: 125189. |
| [78] | MARTIKAINEN PJ. Heterotrophic nitrification-an eternal mystery in the nitrogen cycle[J]. Soil Biology and Biochemistry, 2022, 168: 108611 DOI:10.1016/j.soilbio.2022.108611. |
| [79] | HE TX, XIE DT, NI JP, LI Z, LI ZL. Characteristics of nitrogen transformation and intracellular nitrite accumulation by the hypothermia bacterium Arthrobacter arilaitensis[J]. Science of the Total Environment, 2020, 701: 134730 DOI:10.1016/j.scitotenv.2019.134730. |
| [80] | LIU HY, DING Y, ZHANG QC, LIU XM, XU JM, LI Y, DI HJ. Heterotrophic nitrification and denitrification are the main sources of nitrous oxide in two paddy soils[J]. Plant and Soil, 2019, 445(1): 39-53. |
| [81] | ZHANG Y, ZHAO W, CAI Z, MÜLLER C, ZHANG J. Heterotrophic nitrification is responsible for large rates of N2O emission from subtropical acid forest soil in China[J]. European Journal of Soil Science, 2018, 69(4): 646-654 DOI:10.1111/ejss.12557. |
| [82] | FANXP, YIN C, CHEN H, YE MJ, ZHAO YH, LI TQ, WAKELIN SA, LIANG YC. The efficacy of 3, 4-dimethylpyrazole phosphate on N2O emissions is linked to niche differentiation of ammonia oxidizing archaea and bacteria across four arable soils[J]. Soil Biology and Biochemistry, 2019, 130: 82-93 DOI:10.1016/j.soilbio.2018.11.027. |
| [83] | RUSER R, SCHULZ R. The effect of nitrification inhibitors on the nitrous oxide (N2O) release from agricultural soils—a review[J]. Journal of Plant Nutrition and Soil Science, 2015, 178(2): 171-188 DOI:10.1002/jpln.201400251. |
| [84] | ZHAO B, HE YL, HUGHES J, ZHANG XF. Heterotrophic nitrogen removal by a newly isolated Acinetobacter calcoaceticus HNR[J]. Bioresource Technology, 2010, 101(14): 5194-5200 DOI:10.1016/j.biortech.2010.02.043. |
| [85] | XU SY, WU XL, LU HJ. Overlooked nitrogen-cycling microorganisms in biological wastewater treatment[J]. Frontiers of Environmental Science & Engineering, 2021, 15(6): 133. |
 2023, Vol. 63
2023, Vol. 63