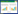中国科学院微生物研究所,中国微生物学会
文章信息
- 石金铭, 陈秋羽, 刘昭曦, 刘双江, 陈敏. 2023
- SHI Jinming, CHEN Qiuyu, LIU Zhaoxi, LIU Shuangjiang, CHEN Min.
- 乳杆菌胞外多糖合成基因簇及构效关系
- Lactobacillus exopolysaccharide: gene clusters for synthesis and structure-activity relationship
- 微生物学报, 63(9): 3482-3499
- Acta Microbiologica Sinica, 63(9): 3482-3499
-
文章历史
- 收稿日期:2023-03-31
- 网络出版日期:2023-07-06
人类的肠道中含有高度复杂的微生态系统——肠道菌群[1-2],该微生态系统的稳态对维护人体健康具有极为关键的作用[3-4]。肠道菌群的调控已逐渐成为维护人体健康和预防治疗疾病的新方法[5]。益生菌是肠道菌群的重要组成部分,参与改善人体肠道微生态。作为具有代表性的一类肠道益生菌,肠道乳酸菌分布在多个属,最著名的属是乳酸杆菌、明串珠菌、趾球菌属、乳球菌属和链球菌属[6]。乳杆菌属(Lactobacillus)是乳酸菌中种数最多的属,有100多种,并且乳杆菌属的许多菌株表现出益生菌特性,在全球食品工业益生菌产品中应用广泛[7],并在发酵食品的生产中发挥重要作用,包括日用乳制品如酸奶、发酵蔬菜等生产过程。乳杆菌对人类的胃肠道系统具有健康益处,它们不仅能产生乳酸、芳香族化合物、过氧化氢、细菌素等,还能产生胞外多糖(exopolysaccharide, EPS)。EPS是一种在微生物生长过程中合成并分泌到细胞外的生物聚合物,其分支程度不同,从线性分子到高支化分子不等[8]。目前乳杆菌胞外多糖的开发存在效率低、纯化困难、安全稳定性差等问题,胞外多糖的结构与功能的对应关系不甚明确;对乳杆菌胞外多糖的结构、合成过程及生理活性进行概述,可以为解决该问题提供借鉴依据。本文总结展望近年来乳杆菌合成胞外多糖的遗传、生物学活性和构效关系等方面的研究进展。
1 乳杆菌胞外多糖生物合成相关酶和基因簇 1.1 乳杆菌胞外多糖的合成与释放依据单糖组成不同,乳杆菌胞外多糖可分为均多糖(homopolysaccharides, HoPS)和杂多糖(heteropolysaccharides, HePS)。乳杆菌在胞外合成HoPS[8],包括聚合和释放两个过程。在聚合过程中,果糖酶[9]和葡萄糖蔗糖酶[10]等相关酶将特定底物(如蔗糖)中的单糖转移到生长的多糖链上;聚合后的HoPS链被释放到细胞外环境中[11-12]。与HoPS相比,HePS结构更多样,生物合成过程比较复杂。微生物通常通过4种途径(胞外合成途径、ABC转运体依赖途径、合酶依赖途径和Wzx/Wzy依赖途径[13])来合成胞外多糖。乳球菌属、乳酸杆菌属和链球菌合成HePS的过程多属Wzx/Wzy依赖途径[14]。Wzx、Wzy为HePS合成及释放过程中发挥关键作用的两种酶,Wzx翻转酶将多糖重复单元易位至周质空间或外膜;Wzy外膜聚合蛋白用于重复单元聚合,调节多糖链长。整个Wzx/Wzy依赖性途径可分为2个步骤,细胞质中的中心碳代谢产生活性糖前体(糖核苷酸),多糖的组装与聚合。Wzx/Wzy依赖性途径合成多糖是一个复杂的细胞内过程,该途径最初发现于革兰氏阴性菌脂多糖(lipopolysaccharide, LPS)的O-抗原多糖的合成过程[15],后在革兰氏阴性菌和革兰氏阳性菌的胞外多糖及荚膜多糖的合成过程中也发现了Wzx/Wzy依赖性途径[13]。该合成途径(图 1)大致分为5步:(1) 单糖/双糖的转运及磷酸化;(2) 单糖活化——糖核苷酸形成;(3) 重复单元合成;(4) 重复单元由细胞膜内表面转运至外表面;(5) 重复单元聚合,长链释放。
|
| 图 1 乳杆菌胞外多糖合成基因簇[20-28] Figure 1 Lactobacillus extracellular polysaccharide synthesis gene cluster[20-28]. A:不同功能的基因由不同颜色的箭头进行注释. B:Wzx/Wzy依赖途径合成胞外多糖过程. 不同的糖底物由不同颜色的圆形表示,Wzx及Wzy由孔道及椭圆表示,糖基转移酶由月牙形表示 A: Genes with different functions are annotated with arrows of different colors. B: Synthesis of extracellular polysaccharides through the Wzx/Wzy pathway. Different sugar substrates are represented by circles of different colors, Wzx and Wzy are represented by pore and elliptical, while glycosyltransferase is represented by a crescent. |
1.2 乳杆菌胞外多糖生物合成基因簇
乳杆菌胞外多糖由EPS基因簇调控合成, 该基因簇通常位于染色质或质粒上[16]。本实验室曾在综述中指出[17]乳酸乳球菌(Lactococcus lactis)和干酪乳酸菌(Lactobacillus casei)等EPS基因簇通常位于质粒上,而嗜热链球菌(Stretococcus thermophilus) Sfi6的EPS基因簇则位于染色体上。HePS中Wzx/Wzy依赖途径前两步涉及的酶通常处在EPS基因簇以外。值得注意的是,同一物种的不同菌株可能具有不同的EPS基因簇,因而具有不同的EPS特征[18]。一般来说,EPS基因簇组织在一个11−22 kb的操纵子中[19],由13−23个特定的功能区组成。但也在部分菌株中发现了多个EPS合成基因簇[20]。已有报道的乳杆菌胞外多糖合成基因簇中,必要基因包括转录调控因子、糖基转移酶(glycosyltransferase, GT)、多糖聚合酶(wzy)和翻转酶(wzx)等。其他基因如LytR转录调节基因(epsA)、磷酸酪氨酸磷酸酶基因(epsD)等常作为EPS合成基因簇的一部分,参与EPS的前体合成和化学装饰。EPS的组成差异多源于糖基转移酶家族的数量和组成不同,见图 1,列举了目前已知的部分乳杆菌胞外多糖合成基因簇[20-28]。
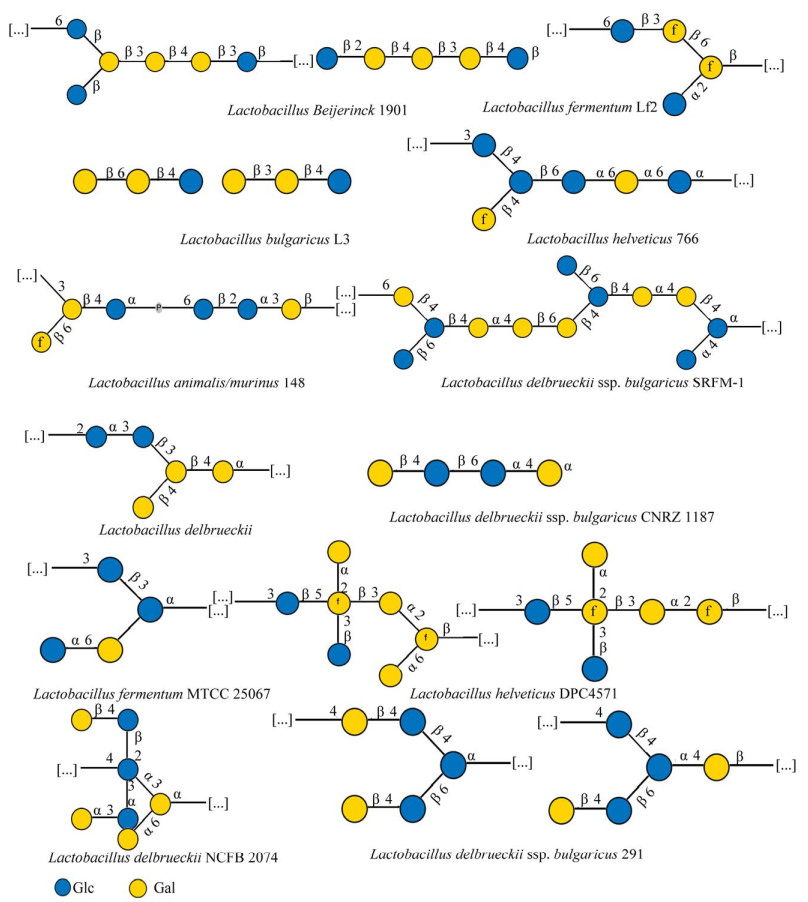
2 乳杆菌胞外多糖的结构和组成乳杆菌胞外多糖的结构可分为一级结构和高级结构。多糖的一级结构是指糖基的组成、排列顺序、相邻糖基的连接方式、糖链有无分支和分支的位置与长短等。糖基上可连接一些官能团如磷酸基团、硫酸基团、甲基化基团,这使得多糖的一级结构更为复杂。多糖的高级结构是在一级结构的基础上,各侧链通过非共价键相互作用结合而形成复杂结构[29],可以理解为分子尺寸。分子生物学及物理化学新技术手段的涌现,为乳杆菌胞外多糖的结构解析提供了新的研究方法[30]。例如一级结构的解析除了常规的化学分析方法,还可利用凝胶柱层析、排阻色谱、气相色谱-质谱联用(gas chromatograph-mass spectrometer, GC-MS)、傅里叶变换红外光谱(Fourier transform infrared spectroscopy, FT-IR)以及核磁共振(nuclear magnetic resonance, NMR)等[31]。高级结构分析的常用方法包括X射线衍射法(X-ray diffraction, XRD)、原子力显微镜法(atomic force microscopy, AFM)与基于高分子稀溶液理论的联用技术、圆二色谱法(circular dichromatography, CD)等[32]。
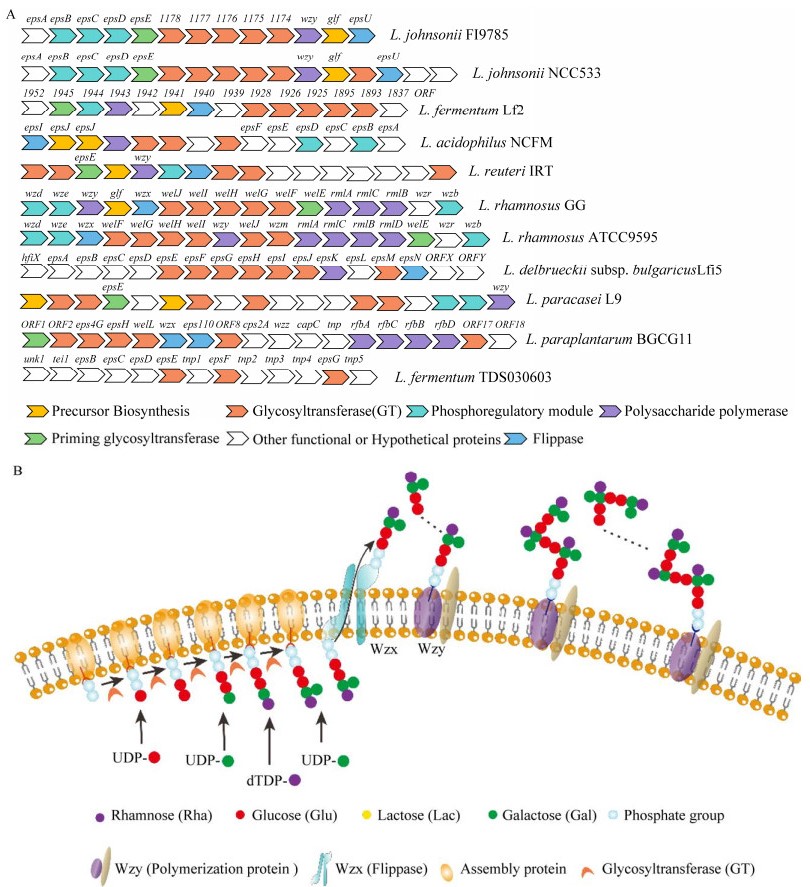
乳杆菌HoPS通常只有糖苷键类型和分子聚合物大小的差异,这归因于细胞外合成依赖途径中合成基因编码的酶的唯一性。根据糖基类型、连接组成和碳键位置,HoPS可分为几种类型,包括d-α-葡聚糖、d-β-葡聚糖、果聚糖和半乳聚糖等[33-34]。乳杆菌HoPS分子量约在4.0×104−6.0×106之间[35]。HoPS主要的聚糖重复单元结构见表 1及图 2[36-49],包括由葡萄糖、半乳糖及果糖组成的糖链结构及键型。
| EPS | Strain | Molecular weight | Linkage | References |
| α-d-glucan | Lactobacillus johnsonii FI9785 | Unknown | α-d-Glcp(1→2)/(1→6) | [36] |
| Lactobacillus. plantarum CIDCA 8327 | Unknown | α-d-Glcp(1→3)/(1→4) | [37] | |
| Lactobacillus brevis E25 | Unknown | α-d-Glcp (1→6) | [38] | |
| Lactobacillus sakei MN1 | 1.74×108 Da | α-d-Glcp (1→6) | [39] | |
| Glucan | Lactobacillus diolivorans G-77 | Unknown | α-d-Glcp(1→2)/β-D-Glcp(1→6) | [40] |
| β-d-glucan | Lactobacillus suebicus CUPV221 | 104−107 Da | β-d-Glcp(1→3) | [41] |
| Lactobacillus brevis TMW 1.2112 | Unknown | β-d-Glcp(1→3)/(1→2) | [42] | |
| Lactobacillus diolivorans G-77, Lactobacillus ethanolidurans CUPV141 | Unknown | β-d-Glcp(1→2)/(1→3) | [40] | |
| Fructan | Lactobacillus reuteri strain 121 | 1.5×105 & 2×106 Da | β-d-Fruf(2→6) | [43] |
| Lactobacillus. reuteri strain 121 | > 107 Da | β-d-Fruf(2→1) | [44] | |
| Lactobacillus reuteri LTH 5448, Lactobacillus sanfranciscensis LTH 2590 | Unknown | β-d-Fruf(1→6) | [45] | |
| Mannan Galactan |
Lactobacillus crispatus L1 | Unknown | α-d-Manp-(1→2)/(1→6)/(1→3) | [36] |
| Lactobacillus mucosae VG1 | Unknown | α-d-Gal(1→6)/β-D-Gal(1→3)/(1→6) | [46] |
|
| 图 2 乳杆菌中常见的均多糖聚糖重复单元结构[49] Figure 2 Common homoglycan repeat unit structures in Lactobacillus[49]. Blue circle represents glucose, yellow circle represents galactose, green circle represents mannose, and green pentagon represents fructose. The shaded portion is a repeat of the partial structure in the glycan unit structure. |
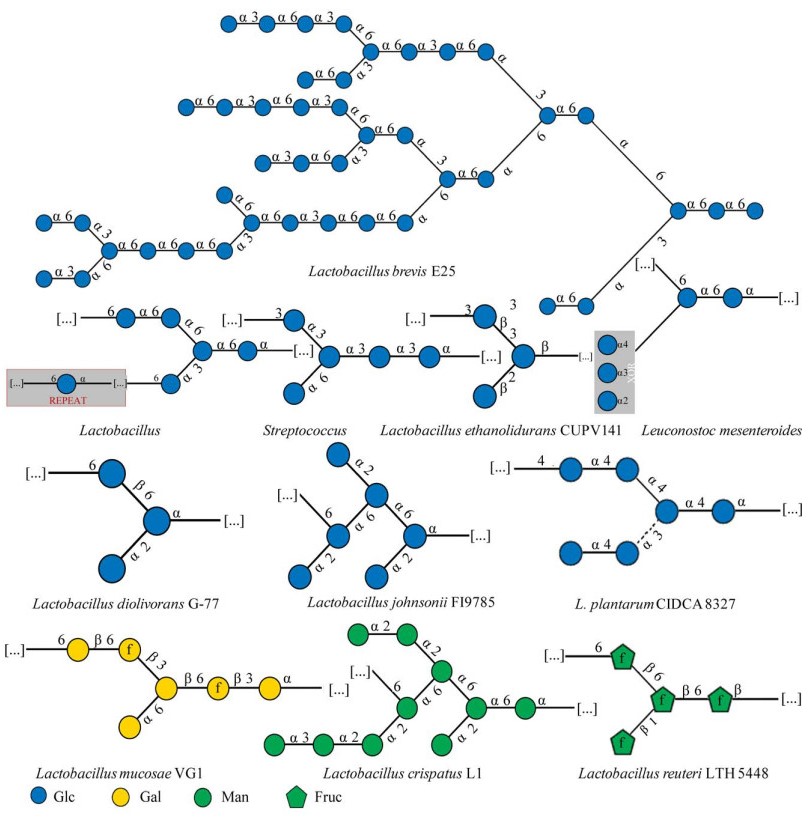
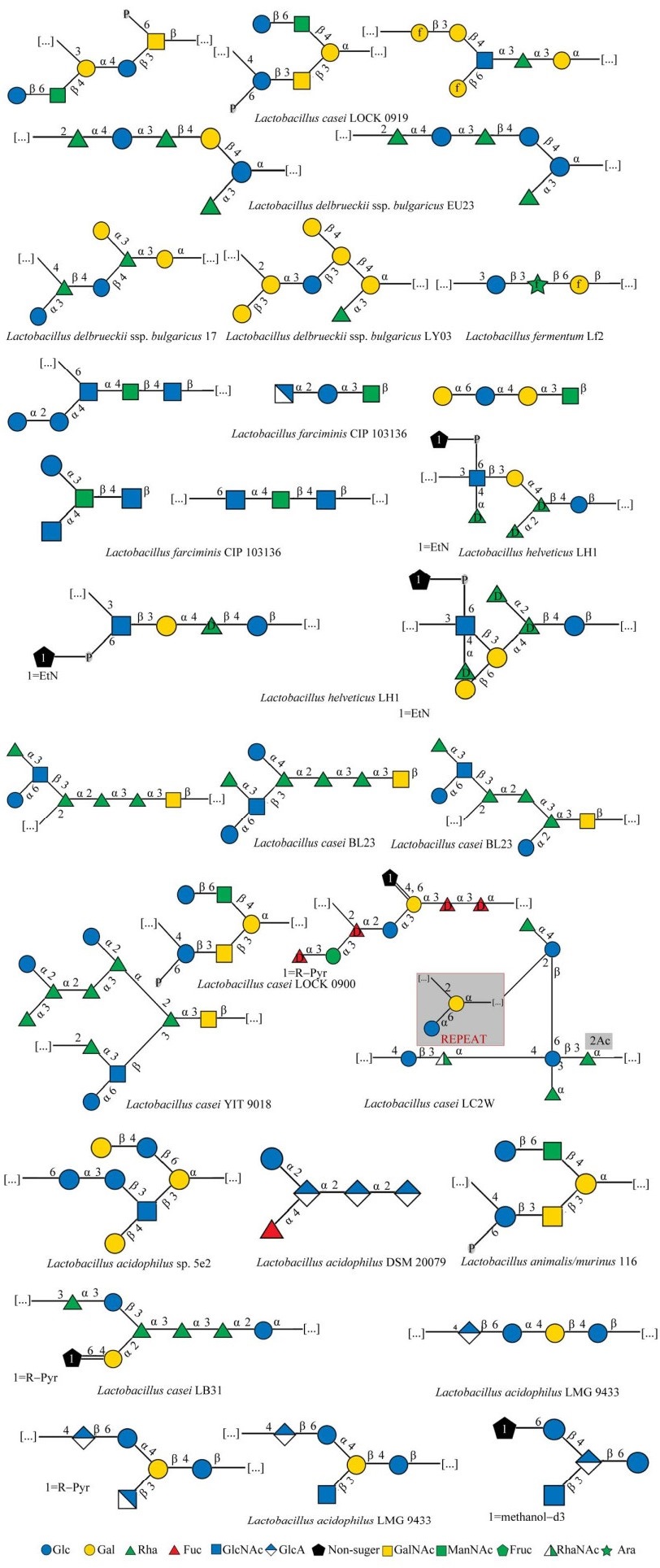
乳杆菌HePS基因簇的差异性决定了HePS生物合成步骤的复杂性[47]。HePS生物合成位置以及Wzx/Wzy依赖途径中底物与酶的多样性,是造成HePS结构多样性的主要原因[38]。一方面,糖链长度与HePS的分子量和分支程度有关。另一方面,重复单元是由多种糖基转移酶调控合成的[48],因此HePS聚合物具有从分子组成到框架键型的巨大结构多样性。中性EPS由两种或两种以上不同的中性单糖组成,而酸性EPS的组成比中性EPS更复杂,还含有一至多个醛酸单糖。此外,磺化和磷酸化等酸性取代基也会增加HePS的酸度,而乙酰基等中性取代基则不会影响到酸碱度的变化。HePS最常见的单糖组成为葡萄糖(Glc)和半乳糖(Gal),多为β型糖苷键。图 3列举了由最常见的2种单糖——葡萄糖和半乳糖组成的聚糖重复单元结构[49]。另外,3种及3种以上单糖组成的聚糖重复单元部分结构见图 4[49],主要有岩藻糖(fucose, Fuc)、甘露糖(mannose, Man)、N-乙酰半乳糖胺(N-acetylgalactosamine, GalNAc)、葡糖胺(glucosamine, GlcN)等作为聚合物的组分。
|
| 图 4 乳杆菌中杂多糖聚糖重复单元结构(以3种单糖及以上为基本单糖组成的HePS)[49] Figure 4 Structure of heteropolysaccharide glycan repeating unit in Lactobacillus[49]. HePS, which is composed of three or more monosaccharides as basic monosaccharides, uses green, red and white green triangles to represent rhamnose, fucose and N-acetylrhamnosamine, blue square to represent N-acetylglucosamine, blue white diamond to represent glucosamine, yellow square to represent N-acetyl galactose amine, green square to represent N-acetylmannosamine, and green star to represent arabinose. The shaded portion is a repeat of the partial structure in the glycan unit structure. Other illustrations are the same as figure 2. |
3 乳杆菌胞外多糖构效关系
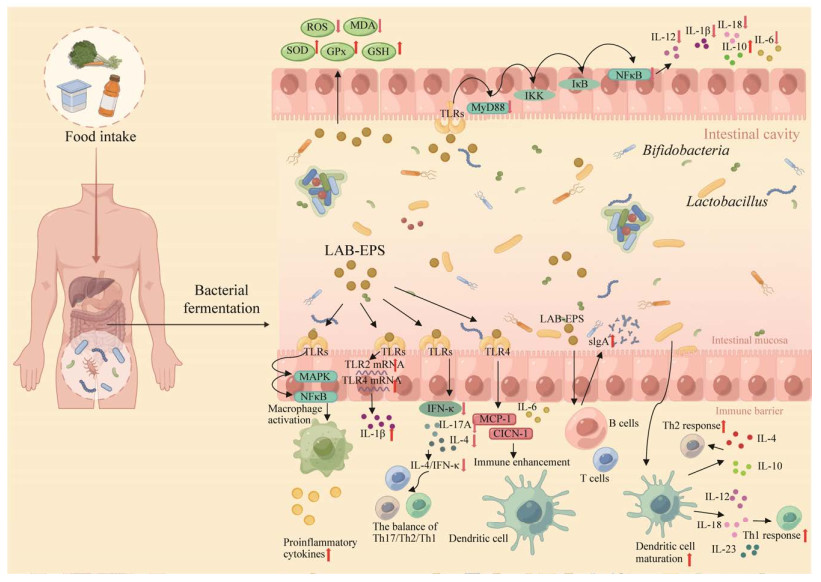
共生肠道微生物群对宿主的代谢和防御系统都有影响[50-51]。益生元如低聚半乳糖(galacto-oligosaccharides, GOS)[52-53]被广泛用于调节肠道菌群功能和丰度。乳杆菌胞外多糖的特异性免疫和非特异性免疫反应通路已经有多个实验验证[54]。几种宿主传感器[55]及其相关的信号通路[56]也已得到阐明。对乳杆菌益生菌活性的效应机制可以聚焦于肠-X轴,例如有研究者称来自植物乳杆菌HY7714的胞外多糖通过皮肤-肠道轴通讯防止皮肤老化[57]。用EPS治疗癌症细胞系,是通过抑制相应基因的表达,导致核转录因子-κB (nuclear factor-kappa B, NF-κB)下调或失活[58]。简述了部分肠道微生态中乳杆菌免疫调节及修复肠道屏障的信号通路,乳杆菌多糖可以通过酶促和非酶促抗氧化系统的联合作用防止活性氧(reactive oxygen species, ROS)的代谢产物对肠上皮的损伤;还可以抑制促炎因子的表达,如IL-1β、IL-6和TNF-α,这些因子是在激活树突状细胞和巨噬细胞的过程中产生的,以响应共生微生物群和TLR信号转导。乳杆菌胞外多糖具有抗癌和抗氧化活性[59],具有抑菌、降低胆固醇、抑制α-淀粉酶和癌细胞系的特性[60]。乳杆菌胞外多糖的抗癌活性与其抗增殖和促凋亡特性有关[61],例如刺激免疫细胞(主要是T和B淋巴细胞、巨噬细胞和NK细胞)释放白细胞介素(图 5)[62]。EPS在各种癌症细胞系中具有剂量和时间依赖性的抗增殖作用。这些性质与其单糖组成、分子量、官能团、聚合主链和侧链结构以及分支点数量有关[63-66],下面将综合论述。
|
| 图 5 肠道稳态中乳杆菌胞外多糖的部分调节作用[58, 62] Figure 5 Partial regulatory effects of Lactobacillus extracellular polysaccharides in intestinal homeostasis[58, 62]. The extracellular polysaccharides of Lactobacillus activate a series of signaling reactions by stimulating TLR receptors, regulating the secretion of various inflammatory signaling factors, thereby affecting the immune response of various immune cells (dendritic cells, B cells, T cells), and regulating intestinal homeostasis. Using different colored balls to represent various inflammatory factors, and brown balls to represent Lactobacillus extracellular polysaccharides. |
3.1 乳杆菌胞外多糖主链糖苷键构效关系
在种类繁多的乳杆菌中,胞外多糖糖苷键对其活性影响较大。如乳杆菌胞外多糖中的1, 4-半乳糖或1, 4-葡萄糖在生物活性中发挥着关键作用。部分研究成果表明,主链结构中含有1, 4-半乳糖或1, 4-葡萄糖的胞外多糖具有更强的抗肿瘤活性[34]。Di等对干酪乳杆菌L. casei SB27两种多糖进行结构鉴定,发现2种多糖主链均具1, 4-半乳糖或1, 4-葡萄糖结构,且对结肠癌抑制率可以达到77.19%和70.87%[67];Li等对瑞士乳杆菌MB2-1结构的研究表明,瑞士乳杆菌MB2-1的EPS组分一的主链含有1, 4-半乳糖和1, 4-葡萄糖,并且该EPS对结肠癌细胞Caco-2的增殖有显著的抑制效果[68]。
乳杆菌胞外多糖的活性与糖链上糖苷键构象有关。β-葡聚糖是具有生物活性的多糖,最经典的结构是以β-1, 3糖苷键为主链,且含有β-1, 6糖苷键连接的支链。β-糖苷键比α-糖苷键构成的葡聚糖的抗肿瘤活性高[63]。L. lactis subsp. cremoris B40菌株产生的EPS中,由β-1, 4糖苷键连接比β-1, 3或α-1, 4糖苷键连接具有更强的刚性,并且α型键比β型键构成的链更具有柔性,进而影响多糖的黏度[69]。Haroun等[70]从L. plantarum NRRL B-4496中分离得到一种以β-1, 4和β-1, 6糖苷键连接而成的葡聚糖,发现它在体外可以抑制6种癌细胞。刘宇等[71]从保加利亚乳杆菌OLL1073R-1分离得到EPS-B16,发现糖苷键较少的酸性多糖能够有效的促进小鼠淋巴细胞增殖,具有抗肿瘤活性。刘滔[72]分离出的植物乳杆菌L. plantarum HY的EPS的糖环全为吡喃糖环,主要为α构型,且存在α-d-甘露糖和α-d-葡萄糖残基,HY-EPS能够还原Fe3+和Mo6+,具有体外抗氧化能力,对α-淀粉酶的活性具有抑制作用,并且具有降血糖潜力。
对乳杆菌胞外多糖的高级结构与抗肿瘤活性之间的构效关系研究可以植物多糖作为参考[63, 73-75],三股螺旋构型是多糖最具活性的空间构象,与α型相比,β螺旋形立体结构由于能形成三股绳状螺旋立体构型,因此抗肿瘤活性更高。目前普遍认为多糖链上以β-1, 6主链或β-1, 3主链葡聚糖时具有较好的抗肿瘤活性[76]。
3.2 乳杆菌胞外多糖分子量构效关系乳杆菌胞外多糖的活性与多糖相对分子质量有关。Asker等[77]表明需土乳杆菌的EPS分子量越小,抗氧化能力越强,推测是因为低分子量的多糖更易结合自由基。在Lactococcus lactis subsp. cremoris B40菌株产生的EPS中,黏度与多糖分子量增加相关[69]。相对分子质量较小的多糖,无法形成有活性的复杂的高级聚合结构,而相对分子质量高的多糖分子体积大,不利于多糖跨膜运输进入生物体内发挥作用[78]。研究推测,小分子量和负电荷的EPS是强免疫调节剂,而分子量较大的中性聚合物是弱免疫调节剂,具有免疫抑制活性[79]。对L. confuses TISTR 1498[80]合成的EPS (5.06×108 g/mol))进行降解,发现EPS (2.9×104 g/mol)更能显著促进巨噬细胞分泌NO,并可提高多种细胞因子(iNOS、IL-10、IL-6和IL-1β)的mRNA表达水平。这是由于低分子量EPS与细胞受体之间具有更好的结合能力,从而促进细胞因子的表达和分泌。L. casei LC2W[81]合成的2种中性EPS组分中,分子质量较低的组分(2.14×104 Da)能够更好地促进小鼠T淋巴细胞增殖。
3.3 乳杆菌胞外多糖单糖摩尔比构效关系对3株植物乳杆菌L. plantarum SKTl 09、L. plantarum Ywl 1和L. plantarum Yw 32[36]所合成EPS进行抗癌实验发现,L. plantarum Ywl 1 EPS (单糖组成摩尔比Glc: Gal=2.71:1)对人结肠癌细胞HT-29细胞的抑制率高于另外2种菌株的EPS (单糖组成摩尔比Fru: Glc=3:1和Man: Fru: Gal: Gal: Glc=8.2:1.0:4.1:4.2)。酸性单糖的含量对多糖的体外抗氧化能力具有重要影响。瑞士乳杆菌MB2-1[82]3种胞外多糖体外抗氧化能力的顺序依次为LHEPS-3 > LHEPS-2 > LHEPS-1,均是由甘露糖、葡萄糖和半乳糖组成,其各自单糖组成的摩尔比为1:2.75:1.33、9.34:1.43:1和2.96:1:1.17;中性糖含量分别为97.85%、94.54%和67.62%;糖醛酸含量分别为0.53%、1.96%和2.53%;硫酸基团含量分别为0.27%、0.42%和0.87%。瑞士乳杆菌SNA12[83]中分离的胞外多糖(SNA12-EPS),富含半乳糖和葡萄糖,摩尔比为1.1:0.12,通过人体粪便发酵的体外模拟消化实验,证明其可提高肠道微生物群产生短链脂肪酸的能力。L. rhamnosus发酵产生的NCVP-F新型α-吡喃多糖有较高的甘露糖和葡萄糖醛酸摩尔比,通过提高抗氧化酶活性(SOD、GSH和GSH-Px),抑制细胞因子水平(IL-6、IL-1β、TNF-α和IL-18),可以更有效地减轻镉损伤小鼠模型的肝肾损伤。在过去10年中,口服益生菌,例如嗜酸乳杆菌(L. acidophilus)、双歧杆菌(B. bifidum)、阿克曼菌(Akkermansia spp.)、丙酸杆菌(Propionibacterium spp.)和益生元补充剂(例如甘露糖、半乳糖、果糖、木糖、异麦芽糖和乳果糖的低聚物)已被用于促进肠道益生菌在结肠中的黏附和定植,缓和肠道微生物的生态失调[84-85]。
3.4 乳杆菌胞外多糖主链分支构效关系LAB-EPS的活性与主链上分支点有关。活性最强的多糖分支度一般在0.20−0.33[86]。有研究者提出相对分子质量较高的多糖形成三螺旋结构是需要β-d-吡喃葡萄糖基分支侧链来提高稳定性的[87]。已有研究证明,乳杆菌可以通过刺激抗癌作用来调节和缓解癌症,例如,EPS结构中甘露糖和葡萄糖残基以及重复单元分支点的存在会增加它们的抗癌活性[68];再如增强癌细胞的凋亡和保护它们免受氧化应激的影响[88]。
3.5 乳杆菌胞外多糖糖链基团及糖链修饰构效关系乳杆菌胞外多糖的抗肿瘤活性随着多糖支链中的羟基数目的增多而增强,这与糖链上存在的羟基与氧自由基之间存在相互作用密切相关,同时随着糖链上羟基数目的增多,其抗氧化活性增强[89-90]。
研究表明,经过羧甲基、硫酸基、乙酰基、苯甲基及磷酸基等化学修饰后的胞外多糖的生物学活性可以得到较大幅度的提高[91]。Tsiapali等[92]对葡聚糖及其衍生物的抗氧化活性进行了测定,发现磷酸化和硫酸化后的葡聚糖抗氧化能力更强,并且硫酸化程度越高,抗氧化活性越强。Li等[66]对Streptococcus thermophilus ASCC 1275胞外多糖进行硫酸化修饰,通过超氧化物和羟基自由基清除试验及Fe3+还原试验对硫酸化EPS的抗氧化活性进行测定,结果表明, 硫酸化修饰EPS的抗氧化活性显著提高(P < 0.05)。乳酸菌可以与病毒直接相互作用、产生抑制物质或刺激免疫等来发挥抗病毒活性[93]。硫酸化多糖即使在粗形式下也具有抗病毒作用[94]。从发酵蔬菜中分离出的L. plantarum IMB19[95],其荚膜多糖富含鼠李糖,为线性九糖重复单元,并由α-葡萄糖和丙氨酸进行修饰;在体外脾细胞培养系统中,该荚膜多糖可增强免疫刺激性细胞因子(IFNγ、TNF-α、IL-6和IL-12)或免疫调节性(IL-10)细胞因子。Zhao等[96]将多糖硒化修饰后,与未修饰的多糖分别作为抗氧化剂,探究对DPPH、羟基自由基和超氧自由基的清除能力,发现硒化修饰的多糖可以显著增强抗氧化的活性,这也是多糖化学改性的实例[97]。刘鹭等[98]研究了硒化修饰后EPS对小鼠腹腔巨噬细胞游离Ca2+的影响,结果表明Se-EPS能够显著提高巨噬细胞中游离Ca2+的浓度,增加巨噬细胞被激活的机会, 分泌更多的细胞因子,进而发挥其免疫细胞的功能。乳杆菌胞外多糖具有机体免疫调节作用。对L. plantarum KF 5[99]发酵合成的EPS单糖组成进行分析,依据T淋巴细胞增殖实验结果,发现有氨基基团且单糖种类多的胞外多糖免疫调节活性更高。另有研究发现,乳杆菌合成的酸性EPS中,磷酸取代基能有效提高巨噬细胞的活性,使抗体的柄端能更有效的与吞噬细胞表面受体进行结合,进而被吞噬;而这些是中性EPS所不能实现的[100]。
4 总结与展望乳杆菌胞外多糖能够通过调节人体免疫系统从而增强机体的抗逆作用,是一种天然的提高机体免疫力的活性物质。可广泛应用在益生保健食品及药品领域中。本实验室曾对肠道共生菌的组成、利用进入肠道内多糖的机制以及产生的代谢产物可能对人体健康存在的潜在影响等方面进行过综述。帮助深入了解肠道菌群的组成结构以及多糖代谢机制,以便未来可以通过某些特定多糖或益生元来精准调控肠道多糖[101]。
由于技术限制,乳杆菌胞外多糖的产量仍不尽如人意。如何提高产量及安全稳定性是急需解决的问题,乳杆菌胞外多糖结构及功能的差异意味着很难建立一个通用的生产流程及标准。
随着糖化学、糖组学、高分辨质谱技术等前沿科学的快速发展,多糖科学迎来了技术发展的新浪潮,也加速推动了多糖微生物学在医药、保健食品和生物材料等领域的应用。微生物胞外多糖介导许多重要的生物过程,如乳杆菌调节人类肠道稳态的作用等。利用细菌表面多糖来研发疫苗和益生制剂已经屡见不鲜。由于微生物多糖结构高度的复杂性和可变性,对其进行系统研究,找出细菌表面多糖共有糖结构仍然具有挑战性。在本实验室相关研究中[102],对CSDB (碳水化合物结构数据库)中的所有微生物多糖结构进行筛选,并分析了所有亚结构的出现次数和物种分布数量。结果表明,不同微生物中存在共同的多糖亚结构。进一步分析表明,这些多糖亚结构可能与细菌的种类、致病性和进化有关。总的来说,这为发现隐藏的信息和聚糖的生物学功能提供了一种替代方法或线索。
多糖的发展还有很长的路要走。在目前的技术水平下,多糖的结构表征和分析仍然是一项极具挑战性的任务。虽然目前对乳杆菌胞外多糖的遗传研究,单糖组成及结构,生理活性及免疫特性的研究已经取得一定的进展。对于乳杆菌EPS的一级结构及构效关系的研究已较为明确,但对其提高机体抗肿瘤和免疫能力的高级结构特征及构效机制的研究还存在明显的不足,尚未形成构效从低级到高级的系统性框架。所以今后应侧重对乳杆菌胞外多糖结构特性与功能特性的系统性研究,从而使其能够在人体共生菌群稳态领域中得到进一步的发展和应用。
| [1] | XU XF, XU PP, MA C, TANG J, ZHANG XW. Gut microbiota, host health, and polysaccharides[J]. Biotechnology Advances, 2013, 31(2): 318-337 DOI:10.1016/j.biotechadv.2012.12.009. |
| [2] | SAA P, URRUTIA A, SILVA-ANDRADE C, MARTÍN AJ, GARRIDO D. Modeling approaches for probing cross-feeding interactions in the human gut microbiome[J]. Computational and Structural Biotechnology Journal, 2021, 20: 79-89. |
| [3] | LEVY M, BLACHER E, ELINAV E. Microbiome, metabolites and host immunity[J]. Current Opinion in Microbiology, 2017, 35: 8-15 DOI:10.1016/j.mib.2016.10.003. |
| [4] | ALTAMIRANO Á, SAA PA, GARRIDO D. Inferring composition and function of the human gut microbiome in time and space: a review of genome-scale metabolic modelling tools[J]. Computational and Structural Biotechnology Journal, 2020, 18: 3897-3904 DOI:10.1016/j.csbj.2020.11.035. |
| [5] | LAVELLE A, SOKOL H. Gut microbiota-derived metabolites as key actors in inflammatory bowel disease[J]. Nature Reviews Gastroenterology & Hepatology, 2020, 17(4): 223-237. |
| [6] | GARBACZ K. Anticancer activity of lactic acid bacteria[J]. Seminars in Cancer Biology, 2022, 86: 356-366 DOI:10.1016/j.semcancer.2021.12.013. |
| [7] | OLEKSY M, KLEWICKA E. Exopolysaccharides produced by Lactobacillus sp. : biosynthesis and applications[J]. Critical Reviews in Food Science and Nutrition, 2018, 58(3): 450-462. |
| [8] | WELMAN AD, MADDOX IS. Exopolysaccharides from lactic acid bacteria: perspectives and challenges[J]. Trends in Biotechnology, 2003, 21(6): 269-274 DOI:10.1016/S0167-7799(03)00107-0. |
| [9] | ANWAR MA, KRALJ S, PIQUÉ AV, LEEMHUIS H, van der MAAREL MJEC, DIJKHUIZEN L. Inulin and levan synthesis by probiotic Lactobacillus gasseri strains: characterization of three novel fructansucrase enzymes and their fructan products[J]. Microbiology (Reading, England), 2010, 156(Pt 4): 1264-1274. |
| [10] | CÔTÉ GL, SKORY CD. Cloning, expression, and characterization of an insoluble glucan-producing glucansucrase from Leuconostoc mesenteroides NRRL B-1118[J]. Applied Microbiology and Biotechnology, 2012, 93(6): 2387-2394 DOI:10.1007/s00253-011-3562-2. |
| [11] | GUPTA P, DIWAN B. Bacterial exopolysaccharide mediated heavy metal removal: a review on biosynthesis, mechanism and remediation strategies[J]. Biotechnology Reports, 2017, 13: 58-71 DOI:10.1016/j.btre.2016.12.006. |
| [12] | ZANNINI E, WATERS DM, COFFEY A, ARENDT EK. Production, properties, and industrial food application of lactic acid bacteria-derived exopolysaccharides[J]. Applied Microbiology and Biotechnology, 2016, 100(3): 1121-1135 DOI:10.1007/s00253-015-7172-2. |
| [13] | SCHMID J, SIEBER V, REHM B. Bacterial exopolysaccharides: biosynthesis pathways and engineering strategies[J]. Frontiers in Microbiology, 2015, 6: 496. |
| [14] | ZEIDAN AA, POULSEN VK, JANZEN T, BULDO P, DERKX PMF, ØREGAARD G, NEVES AR. Polysaccharide production by lactic acid bacteria: from genes to industrial applications[J]. FEMS Microbiology Reviews, 2017, 41(supp_1): S168-S200 DOI:10.1093/femsre/fux017. |
| [15] | ISLAM ST, LAM JS. Synthesis of bacterial polysaccharides via the Wzx/Wzy-dependent pathway[J]. Canadian Journal of Microbiology, 2014, 60(11): 697-716 DOI:10.1139/cjm-2014-0595. |
| [16] |
LI M. Structure of extracellular polysaccharide and transcription analysis of eps gene cluster of Lactococcus lactis subsp. lactis IMAU11823[D]. Hohhot: Master's Thesis of Inner Mongolia Agricultural University (in Chinese). 李敏. 乳酸乳球菌乳酸亚种IMAU11823胞外多糖结构及eps基因簇转录分析[D]. 呼和浩特: 内蒙古农业大学硕士学位论文. |
| [17] |
童良琴, 曲亚军, 陈敏. 乳酸菌胞外多糖的研究进展[J]. 中国生物工程杂志, 2015, 35(11): 85-91.
DOI:10.13523/j.cb.20151112 TONG LQ, QU YJ, CHEN M. Research advance on exopolysaccharides synthesized by lactic acid bacteria[J]. China Biotechnology, 2015, 35(11): 85-91 (in Chinese). |
| [18] | ZHOU Y, CUI Y, QU X. Exopolysaccharides of lactic acid bacteria: structure, bioactivity and associations: a review[J]. Carbohydrate Polymers, 2019, 207: 317-332 DOI:10.1016/j.carbpol.2018.11.093. |
| [19] | CUI YH, JIANG X, HAO MY, QU XJ, HU T. New advances in exopolysaccharides production of Streptococcus thermophilus[J]. Archives of Microbiology, 2017, 199(6): 799-809 DOI:10.1007/s00203-017-1366-1. |
| [20] | ALE EC, ROJAS MF, REINHEIMER JA, BINETTI AG. Lactobacillus fermentum: could EPS production ability be responsible for functional properties?[J]. Food Microbiology, 2020, 90: 103465 DOI:10.1016/j.fm.2020.103465. |
| [21] | DAN T, FUKUDA K, SUGAI-BANNAI M, TAKAKUWA N, MOTOSHIMA H, URASHIMA T. Characterization and expression analysis of the exopolysaccharide gene cluster in Lactobacillus fermentum TDS030603[J]. Bioscience, Biotechnology, and Biochemistry, 2009, 73(12): 2656-2664 DOI:10.1271/bbb.90502. |
| [22] | HUANG Z, ZHOU XY, STANTON C, ROSS RP, ZHAO JX, ZHANG H, YANG B, CHEN W. Comparative genomics and specific functional characteristics analysis of Lactobacillus acidophilus[J]. Microorganisms, 2021, 9(9): 1992 DOI:10.3390/microorganisms9091992. |
| [23] | LEBEER S, VERHOEVEN TLA, FRANCIUS G, SCHOOFS G, LAMBRICHTS I, DUFRÊNE Y, VANDERLEYDEN J, de KEERSMAECKER SCJ. Identification of a gene cluster for the biosynthesis of a long, galactose-rich exopolysaccharide in Lactobacillus rhamnosus GG and functional analysis of the priming glycosyltransferase[J]. Applied and Environmental Microbiology, 2009, 75(11): 3554-3563 DOI:10.1128/AEM.02919-08. |
| [24] | PÉANT B, LAPOINTE G, GILBERT C, ATLAN D, WARD P, ROY D. Comparative analysis of the exopolysaccharide biosynthesis gene clusters from four strains of Lactobacillus rhamnosus[J]. Microbiology, 2005, 151(6): 1839-1851 DOI:10.1099/mic.0.27852-0. |
| [25] | VASTANO V, PERRONE F, MARASCO R, SACCO M, MUSCARIELLO L. Transcriptional analysis of exopolysaccharides biosynthesis gene clusters in Lactobacillus plantarum[J]. Archives of Microbiology, 2016, 198(3): 295-300 DOI:10.1007/s00203-015-1169-1. |
| [26] | DEO D, DAVRAY D, KULKARNI R. A diverse repertoire of exopolysaccharide biosynthesis gene clusters in Lactobacillus revealed by comparative analysis in 106 sequenced genomes[J]. Microorganisms, 2019, 7(10): 444 DOI:10.3390/microorganisms7100444. |
| [27] | REMUS DM, van KRANENBURG R, van SWAM II, TAVERNE N, BONGERS RS, WELS M, WELLS JM, BRON PA, KLEEREBEZEM M. Impact of 4 Lactobacillus plantarum capsular polysaccharide clusters on surface glycan composition and host cell signaling[J]. Microbial Cell Factories, 2012, 11(1): 1-10 DOI:10.1186/1475-2859-11-1. |
| [28] | ZIVKOVIC M, MILJKOVIC M, RUAS-MADIEDO P, STRAHINIC I, TOLINACKI M, GOLIC N, KOJIC M. Exopolysaccharide production and ropy phenotype are determined by two gene clusters in putative probiotic strain Lactobacillus paraplantarum BGCG11[J]. Applied and Environmental Microbiology, 2015, 81(4): 1387-1396 DOI:10.1128/AEM.03028-14. |
| [29] |
何余堂, 潘孝明. 植物多糖的结构与活性研究进展[J]. 食品科学, 2010, 31(17): 493-496.
HE YT, PAN XM. Biological activity and structure of plant polysaccharides[J]. Food Science, 2010, 31(17): 493-496 (in Chinese). |
| [30] | SALAMA Y, CHENNAOUI M, SYLLA A, MOUNTADAR M, RIHANI M, ASSOBHEI O. Characterization, structure, and function of extracellular polymeric substances (EPS) of microbial biofilm in biological wastewater treatment systems: a review[J]. Desalination and Water Treatment, 2016, 57(35): 16220-16237 DOI:10.1080/19443994.2015.1077739. |
| [31] | ZHANG Z, Wang F, WANG M, MA L, YE H, ZENG X. A comparative study of the neutral and acidic polysaccharides from Allium macrostemon Bunge[J]. Carbohydrate Polymers, 2015, 117: 980-987 DOI:10.1016/j.carbpol.2014.10.019. |
| [32] | LIU Z, ZHANG Z, QIU L, ZHANG F, XU X, WEI H, TAO X. Characterization and bioactivities of the exopolysaccharide from a probiotic strain of Lactobacillus plantarum WLPL04[J]. Journal of Dairy Science, 2017, 100(9): 6895-6905 DOI:10.3168/jds.2016-11944. |
| [33] | CAGGIANIELLO G, KLEEREBEZEM M, SPANO G. Exopolysaccharides produced by lactic acid bacteria: from health-promoting benefits to stress tolerance mechanisms[J]. Applied Microbiology and Biotechnology, 2016, 100(9): 3877-3886 DOI:10.1007/s00253-016-7471-2. |
| [34] |
邸维, 张兰威, 易华西, 韩雪. 乳酸菌胞外多糖结构及其功能多样性的研究进展[J]. 中国乳品工业, 2017, 45(5): 32-37.
DI W, ZHANG LW, YI HX, HAN X. Research advances on structure and diversity function of exopolysaccharides produced by lactic acid bacteria[J]. China Dairy Industry, 2017, 45(5): 32-37 (in Chinese). |
| [35] | RUAS-MADIEDO P, HUGENHOLTZ J, ZOON P. An overview of the functionality of exopolysaccharides produced by lactic acid bacteria[J]. International Dairy Journal, 2002, 12(2/3): 163-171. |
| [36] | CASTRO-BRAVO N, WELLS JM, MARGOLLES A, RUAS-MADIEDO P. Interactions of surface exopolysaccharides from Bifidobacterium and Lactobacillus within the intestinal environment[J]. Frontiers in Microbiology, 2018, 9: 2426 DOI:10.3389/fmicb.2018.02426. |
| [37] | GANGOITI MV, PUERTAS AI, HAMET MF, PERUZZO PJ, LLAMAS MG, MEDRANO M, PRIETO A, DUEÑAS MT, ABRAHAM AG. Lactobacillus plantarum CIDCA 8327: an α-glucan producing-strain isolated from kefir grains[J]. Carbohydrate Polymers, 2017, 170: 52-59 DOI:10.1016/j.carbpol.2017.04.053. |
| [38] | DERTLI E, COLQUHOUN IJ, CÔTÉ GL, LE GALL G, NARBAD A. Structural analysis of the α-d-glucan produced by the sourdough isolate Lactobacillus brevis E25[J]. Food Chemistry, 2018, 242: 45-52 DOI:10.1016/j.foodchem.2017.09.017. |
| [39] | ZAROUR K, LLAMAS MG, PRIETO A, RÚAS-MADIEDO P, TERESA DUEÑAS M, de PALENCIA PF, AZNAR R, KIHAL M, LÓPEZ P. Rheology and bioactivity of high molecular weight dextrans synthesised by lactic acid bacteria[J]. Carbohydrate Polymers, 2017, 174: 646-657 DOI:10.1016/j.carbpol.2017.06.113. |
| [40] | KNIREL YA, van CM. Bacterial Exopolysaccharides, in Reference Module in Chemistry, Molecular Sciences and Chemical Engineering[M]. Amsterdam: Elsevier, 2021. |
| [41] | GARAI-IBABE G, TERESA DUEÑAS M, IRASTORZA A, SIERRA-FILARDI E, LAURA WERNING M, LÓPEZ P, CORBÍ AL, FERNÁNDEZ de PALENCIA P. Naturally occurring 2-substituted (1, 3)-β-d-glucan producing Lactobacillus suebicus and Pediococcus parvulus strains with potential utility in the production of functional foods[J]. Bioresource Technology, 2010, 101(23): 9254-9263 DOI:10.1016/j.biortech.2010.07.050. |
| [42] | FRAUNHOFER ME, GEISSLER AJ, WEFERS D, BUNZEL M, JAKOB F, VOGEL RF. Characterization of β-glucan formation by Lactobacillus brevis TMW 1.2112 isolated from slimy spoiled beer[J]. International Journal of Biological Macromolecules, 2018, 107: 874-881 DOI:10.1016/j.ijbiomac.2017.09.063. |
| [43] | van HIJUM SAFT, BONTING K, van der MAAREL MJEC, DIJKHUIZEN L. Purification of a novel fructosyltransferase from Lactobacillus reuteri strain 121 and characterization of the levan produced[J]. FEMS Microbiology Letters, 2001, 205(2): 323-328 DOI:10.1111/j.1574-6968.2001.tb10967.x. |
| [44] | van HIJUM SA, van GEEL-SCHUTTEN GH, RAHAOUI H, van der MAAREL MJEC, DIJKHUIZEN L. Characterization of a novel fructosyltransferase from Lactobacillus reuteri that synthesizes high-molecular-weight inulin and inulin oligosaccharides[J]. Applied and Environmental Microbiology, 2002, 68(9): 4390-4398 DOI:10.1128/AEM.68.9.4390-4398.2002. |
| [45] | SHAHID M, RAJOKA R. Lactobacillus exopolysaccharides: new perspectives on engineering strategies, physiochemical functions, and immunomodulatory effects on host health[J]. Trends in Food Science & Technology, 2020, 103: 36-48. |
| [46] | FAGUNWA O, AHMED HI, SADIQ S, HUMPHREYS PN, MCLAY N, LAWS AP. Isolation and characterization of a novel exopolysaccharide secreted by Lactobacillus mucosae VG1[J]. Carbohydrate Research, 2019, 484: 107781 DOI:10.1016/j.carres.2019.107781. |
| [47] | DU B, YANG Y, BIAN Z, XU B. Molecular weight and helix conformation determine intestinal anti-inflammatory effects of exopolysaccharide from Schizophyllum commune[J]. Carbohydrate Polymers, 2017, 172: 68-77 DOI:10.1016/j.carbpol.2017.05.032. |
| [48] | DU R, XING H, YANG Y, JIANG H, ZHOU Z, HAN Y. Optimization, purification and structural characterization of a dextran produced by L. mesenteroides isolated from Chinese sauerkraut[J]. Carbohydrate Polymers, 2017, 174: 409-416 DOI:10.1016/j.carbpol.2017.06.084. |
| [49] | SCHERBININA SI, FRANK M, TOUKACH PV. Carbohydrate structure database oligosaccharide conformation tool[J]. Glycobiology, 2022, 32(6): 460-468 DOI:10.1093/glycob/cwac011. |
| [50] | DUNCAN PI, AITIO O, HEISKANEN A, NIEMELÄ R, SAARINEN J, HELIN J, PORTA N, FIAUX M, MOËNNOZ D, GOLLIARD M, CHERBUT C, BERROCAL R, AUSTIN S, SPRENGER N. Structure and function of bovine whey derived oligosaccharides showing synbiotic epithelial barrier protective properties[J]. Nutrients, 2020, 12(7): 2007 DOI:10.3390/nu12072007. |
| [51] | GLENDINNING L, FREE A. Supra-organismal interactions in the human intestine[J]. Frontiers in Cellular and Infection Microbiology, 2014, 4: 47. |
| [52] | SODERHOLM AT, PEDICORD VA. Intestinal epithelial cells: at the interface of the microbiota and mucosal immunity[J]. Immunology, 2019, 158(4): 267-280 DOI:10.1111/imm.13117. |
| [53] | OLSZAK T, NEVES JF, DOWDS CM, BAKER K, GLICKMAN J, DAVIDSON NO, LIN CS, JOBIN C, BRAND S, SOTLAR K, WADA K, KATAYAMA K, NAKAJIMA A, MIZUGUCHI H, KAWASAKI K, NAGATA K, MÜLLER W, SNAPPER SB, SCHREIBER S, KASER A, et al. Protective mucosal immunity mediated by epithelial CD1d and IL-10[J]. Nature, 2014, 509(7501): 497-502 DOI:10.1038/nature13150. |
| [54] |
梁增澜, 李超, 王艳萍. 乳酸菌胞外多糖免疫活性的研究进展[J]. 食品与发酵工业, 2018, 44(2): 266-272.
DOI:10.13995/j.cnki.11-1802/ts.015801 LIANG ZL, LI C, WANG YP. Research progressing of immune regulatory activity of exopolysaccharides synthesized by lactic acid bacterium[J]. Food and Fermentation Industries, 2018, 44(2): 266-272 (in Chinese). |
| [55] | CHU H, MAZMANIAN SK. Innate immune recognition of the microbiota promotes host-microbial symbiosis[J]. Nature Immunology, 2013, 14(7): 668-675 DOI:10.1038/ni.2635. |
| [56] | SELLGE G, KUFER TA. PRR-signaling pathways: learning from microbial tactics[J]. Seminars in Immunology, 2015, 27(2): 75-84 DOI:10.1016/j.smim.2015.03.009. |
| [57] | LEE K, KIM HJ, KIM SA, PARK SD, SHIM JJ, LEE JL. Exopolysaccharide from Lactobacillus plantarum HY7714 protects against skin aging through skin-gut axis communication[J]. Molecules, 2021, 26(6): 1651 DOI:10.3390/molecules26061651. |
| [58] | TANIGUCHI K, KARIN M. NF-κB, inflammation, immunity and cancer: coming of age[J]. Nature Reviews Immunology, 2018, 18(5): 309-324 DOI:10.1038/nri.2017.142. |
| [59] | EL GHANY KA, ELHAFEZ EA, HAMOUDA RA, MAHROUS H, ALLAH H AHMED F, HAMZA HA. Evaluation of antioxidant and antitumor activities of Lactobacillus acidophilus bacteria isolated from Egyptian infants[J]. International Journal of Pharmacology, 2014, 10(5): 282-288 DOI:10.3923/ijp.2014.282.288. |
| [60] | DILNA SV, SURYA H, ASWATHY RG, VARSHA KK, SAKTHIKUMAR DN, PANDEY A, NAMPOOTHIRI KM. Characterization of an exopolysaccharide with potential health-benefit properties from a probiotic Lactobacillus plantarum RJF4[J]. LWT-Food Science and Technology, 2015, 64(2): 1179-1186 DOI:10.1016/j.lwt.2015.07.040. |
| [61] | WU J, ZHANG Y, YE L, WANG C. The anti-cancer effects and mechanisms of lactic acid bacteria exopolysaccharides in vitro: a review[J]. Carbohydrate Polymers, 2021, 253: 117308 DOI:10.1016/j.carbpol.2020.117308. |
| [62] | ISMAIL B, NAMPOOTHIRI KM. Molecular characterization of an exopolysaccharide from a probiotic Lactobacillus plantarum MTCC 9510 and its efficacy to improve the texture of starchy food[J]. Journal of Food Science and Technology, 2014, 51(12): 4012-4018 DOI:10.1007/s13197-013-0928-8. |
| [63] |
邱琳, 辛现良, 耿美玉. 多糖构效关系研究进展[J]. 现代生物医学进展, 2009, 9(9): 1764-1768.
QIU L, XIN XL, GENG MY. Advances in the structure-function relationships of polysaccharides[J]. Progress in Modern Biomedicine, 2009, 9(9): 1764-1768 (in Chinese). |
| [64] | HAMURO J, YAMASHITA Y, OHSAKA Y, MAEDA YY, CHIHARA G. Carboxymethylpachymaran, a new water soluble polysaccharide with marked antitumour activity[J]. Nature, 1971, 233(5320): 486-488 DOI:10.1038/233486a0. |
| [65] | YANG JH, DU YM, WEN Y, LI TY, HU L. Sulfation of Chinese lacquer polysaccharides in different solvents[J]. Carbohydrate Polymers, 2003, 52(4): 397-403 DOI:10.1016/S0144-8617(02)00330-2. |
| [66] | LI SQ, SHAH NP. Antioxidant and antibacterial activities of sulphated polysaccharides from Pleurotus eryngii and Streptococcus thermophilus ASCC 1275[J]. Food Chemistry, 2014, 165: 262-270 DOI:10.1016/j.foodchem.2014.05.110. |
| [67] | DI W, ZHANG LW, WANG SM, YI HX, HAN X, FAN RB, ZHANG YC. Physicochemical characterization and antitumour activity of exopolysaccharides produced by Lactobacillus casei SB27 from yak milk[J]. Carbohydrate Polymers, 2017, 171: 307-315 DOI:10.1016/j.carbpol.2017.03.018. |
| [68] | LI W, TANG WZ, JI J, XIA XD, RUI X, CHEN XH, JIANG M, ZHOU JZ, DONG MS. Characterization of a novel polysaccharide with anti-colon cancer activity from Lactobacillus helveticus MB2-1[J]. Carbohydrate Research, 2015, 411: 6-14 DOI:10.1016/j.carres.2014.12.014. |
| [69] | LOOIJESTEIJN PJ, van CASTEREN WHM, TUINIER R, DOESWIJK-VORAGEN CHL, HUGENHOLTZ J. Influence of different substrate limitations on the yield, composition and molecular mass of exopolysaccharides produced by Lactococcus lactis subsp. cremoris in continuous cultures[J]. Journal of Applied Microbiology, 2000, 89(1): 116-122 DOI:10.1046/j.1365-2672.2000.01082.x. |
| [70] | HAROUNB M, REFAAT BM, EL-MENOUFY HA, AMIN HA, EL-WASEIF AA. Structure analysis and antitumor activity of the exopolysaccharide from probiotic Lactobacillus plantarum NRRL B-4496 in vitro and in vivo[J]. Journal of Applied Sciences Research, 2013, 9(1): 425-434. |
| [71] |
刘宇, 孟祥晨. 乳酸菌胞外多糖及其抗肿瘤活性[J]. 中国乳品工业, 2008, 36(1): 39-43.
LIU Y, MENG XC. Expolysaccharides produced by lactic acid bacteria and their anti-tumor activity[J]. China Dairy Industry, 2008, 36(1): 39-43 (in Chinese). |
| [72] |
LIU T. Screening of lactic acid bacteria producing extracellular polysaccharide and study on the structure and in vitro function of polysaccharide[D]. Yaan: Master's Thesis of Sichuan Agricultural University, 2019. (in Chinese). 刘滔. 产胞外多糖乳酸菌的筛选及其多糖的结构和体外功能的研究[D]. 雅安: 四川农业大学硕士学位论文, 2019. |
| [73] |
MIAO F. Study on enzymatic extraction, separation and purification, antioxidant and immune activities of Lycium barbarum polysaccharide[D]. Yangzhou: Master's Thesis of Yangzhou University, 2021 (in Chinese). 缪凤. 枸杞多糖酶提、分离纯化及抗氧化和免疫活性研究[D]. 扬州: 扬州大学硕士学位论文, 2021. |
| [74] |
GUO Q. Extraction, purification, solution properties and structure of Lycium barbarum polysaccharide[D]. Xi'an: Master's Thesis of Shaanxi Normal University 2012 (in Chinese). 郭琦. 枸杞多糖的提取、分离纯化、溶液性质及其结构的初步研究[D]. 西安: 陕西师范大学硕士学位论文, 2012. |
| [75] |
梁小飞, 张芳, 姜胤秀, 刘梦秋, 郭盛, 钱大玮, 段金廒. 枸杞多糖构-效关系研究进展与展望[J]. 中国中药杂志, 2023, 48(9): 2387-2395.
LIANG XF, ZHANG F, JIANG YX, LIU MQ, GUO S, QIAN DW, DUAN JIN'AO. Structure-activity relationship of Lycium barbarum polysaccharides[J]. China Journal of Chinese Materia Medica, 2023, 48(9): 2387-2395 (in Chinese). |
| [76] |
谢渟, 肖春, 王涓, 黄龙花, 吴清平. 灰树花活性多糖构效关系研究进展[J]. 微生物学通报, 2022, 49(8): 3401-3419.
XIE T, XIAO C, WANG J, HUANG LH, WU QP. Advances in structure-activity relationship of polysaccharides from Grifola frondosa[J]. Microbiology China, 2022, 49(8): 3401-3419 (in Chinese). |
| [77] | ASKER MMS, AHMED YM, RAMADAN MF. Chemical characteristics and antioxidant activity of exopolysaccharide fractions from Microbacterium terregens[J]. Carbohydrate Polymers, 2009, 77(3): 563-567 DOI:10.1016/j.carbpol.2009.01.037. |
| [78] |
陈静, 李巧珍, 宋春艳, 章炉军, 尚晓冬, 辜运富. 香菇多糖提取纯化、生物活性及构效关系研究进展[J]. 上海农业学报, 2021, 37(5): 144-150.
CHEN J, LI QZ, SONG CY, ZHANG LJ, SHANG XD, GU YF. Lentinan: a review on the isolation and purification, bioactivities and structure-activity relationship[J]. Acta Agriculturae Shanghai, 2021, 37(5): 144-150 (in Chinese). |
| [79] | SHAO L, WU Z, ZHANG H, CHEN W, AI L, GUO B. Partial characterization and immunostimulatory activity of exopolysaccharides from Lactobacillus rhamnosus KF5[J]. Carbohydrate Polymers, 2014, 107: 51-56 DOI:10.1016/j.carbpol.2014.02.037. |
| [80] | SURAYOT U, WANG JG, SEESURIYACHAN P, KUNTIYA A, TABARSA M, LEE Y, KIM JK, PARK W, YOU S. Exopolysaccharides from lactic acid bacteria: structural analysis, molecular weight effect on immunomodulation[J]. International Journal of Biological Macromolecules, 2014, 68: 233-40 DOI:10.1016/j.ijbiomac.2014.05.005. |
| [81] |
AI LZ. Study on preparation, function and structure of exopolysaccharides from Lactobacillus Casei LC2W[D]. Wuxi: Doctoral Dissertation of Jiangnan University, 2007 (in Chinese). 艾连中. 干酪乳杆菌LC2W胞外多糖制备、功能及结构的研究[D]. 无锡: 江南大学博士学位论文, 2007. |
| [82] |
JI J. Preparation, structure and physiological function of extracellular polysaccharide from Lactobacillus helveticus MB2-1[D]. Nanjing: Master's Thesis of Nanjing Agricultural University, 2014 (in Chinese). 纪鹃. 瑞士乳杆菌MB2-1胞外多糖制备、结构和生理功能的研究[D]. 南京: 南京农业大学硕士学位论文, 2014. |
| [83] | WANG X, XU M, XU D, MA K, ZHANG C, WANG G, DONG M, LI W. Structural and prebiotic activity analysis of the polysaccharide produced by Lactobacillus helveticus SNA12[J]. Carbohydrate Polymers, 2022, 296: 119971 DOI:10.1016/j.carbpol.2022.119971. |
| [84] | ELLEN SANDERS M, MERENSTEIN DJ, REID G, GIBSON GR, RASTALL RA. Probiotics and prebiotics in intestinal health and disease: from biology to the clinic[J]. Nature Reviews Gastroenterology & Hepatology, 2019, 16(10): 605-616. |
| [85] | VERA C, ILLANES A, GUERRERO C. Enzymatic production of prebiotic oligosaccharides[J]. Current Opinion in Food Science, 2021, 37: 160-170 DOI:10.1016/j.cofs.2020.10.013. |
| [86] | MENG X, LIANG H, LUO L. Antitumor polysaccharides from mushrooms: a review on the structural characteristics, antitumor mechanisms and immunomodulating activities[J]. Carbohydrate Research, 2016, 424: 30-41. |
| [87] | SHENG K, WANG C, CHEN B, KANG M, WANG M, LIU K, WANG M. Recent advances in polysaccharides from Lentinus edodes (Berk.): isolation, structures and bioactivities[J]. Food Chemistry, 2021, 358: 129883. |
| [88] | VIVARELLI S, SALEMI R, CANDIDO S, FALZONE L, SANTAGATI M, STEFANI S, TORINO F, BANNA GL, TONINI G, LIBRA M. Gut microbiota and cancer: from pathogenesis to therapy[J]. Cancers, 2019, 11(1): 38. |
| [89] |
索超, 曲晓军, 崔艳华. 乳酸菌胞外多糖研究进展[J]. 中国乳品工业, 2017, 45(11): 32-36.
SUO C, QU XJ, CUI YH. Research advances in extracellular polysaccharide produced by lactic acid bacteria[J]. China Dairy Industry, 2017, 45(11): 32-36 (in Chinese). |
| [90] | YANG J, DU Y, HUANG R, WAN Y, LI T. Chemical modification, characterization and structure-anticoagulant activity relationships of Chinese lacquer polysaccharides[J]. International Journal of Biological Macromolecules, 2002, 31(1/2/3): 55-62. |
| [91] |
LIU J. Study on fermentation conditions, structure, chemical modification and antioxidant activity of paecilomyces polymyxa extracellular polysaccharide[D]. Nanjing: Doctoral Dissertation of Nanjing Agricultural University, 2010 (in Chinese). 刘俊. 多粘类芽孢杆菌胞外多糖的发酵条件、结构、化学修饰及其抗氧化活性的研究[D]. 南京: 南京农业大学博士学位论文, 2010. |
| [92] | TSIAPALI E, WHALEY S, KALBFLEISCH J, ENSLEY HE, BROWDER IW, WILLIAMS DL. Glucans exhibit weak antioxidant activity, but stimulate macrophage free radical activity[J]. Free Radical Biology & Medicine, 2001, 30(4): 393-402. |
| [93] | AL KASSAA I, HOBER D, HAMZE M, CHIHIB NE, DRIDER D. Antiviral potential of lactic acid bacteria and their bacteriocins[J]. Probiotics and Antimicrobial Proteins, 2014, 6(3): 177-185. |
| [94] | LIU J, LUO JG, YE H, SUN Y, LU ZX, ZENG XX. Production, characterization and antioxidant activities in vitro of exopolysaccharides from endophytic bacterium Paenibacillus polymyxa EJS-3[J]. Carbohydrate Polymers, 2009, 78(2): 275-281. |
| [95] | GARCIA-VELLO P, SHARMA G, SPECIALE I, MOLINARO A, IM SH, de CASTRO C. Structural features and immunological perception of the cell surface glycans of Lactobacillus plantarum: a novel rhamnose-rich polysaccharide and teichoic acids[J]. Carbohydrate Polymers, 2020, 233: 115857. |
| [96] | ZHAO BT, ZHANG J, YAO J, SONG S, YIN ZX, GAO QY. Selenylation modification can enhance antioxidant activity of Potentilla anserinab L. polysaccharide[J]. International Journal of Biological Macromolecules, 2013, 58: 320-328. |
| [97] | WANG C, GONG Y, LIN Y, SHEN J, WANG DA. A novel gellan gel-based microcarrier for anchorage-dependent cell delivery[J]. Acta Biomaterialia, 2008, 4(5): 1226-1234. |
| [98] |
刘鹭, 潘道东, 丁琳, 曾小群. 硒化乳酸菌胞外多糖对小鼠腹腔巨噬细胞及肿瘤细胞内游离Ca2+的影响[J]. 食品科学, 2014, 35(1): 250-253.
LIU L, PAN DD, DING L, ZENG XQ. Effect of selenium-modified exopolysaccharide from Lactococcus lactis subsp. lactis on [Ca2+] in mouse macrophages and cancer cells[J]. Food Science, 2014, 35(1): 250-253 (in Chinese). |
| [99] |
SHAO L. Screening of exopolysaccharide-producing Lactobacilli and separation, structure and bioactivities of exopolysaccharide[D]. Wuxi: Doctoral Dissertation of Jiangnan University, 2014 (in Chinese). 邵丽. 产胞外多糖乳杆菌的筛选及其多糖的分离、结构和生物活性研究[D]. 无锡: 江南大学博士学位论文, 2014. |
| [100] | KITAZAWA H, ISHII Y, UEMURA J, KAWAI Y, SAITO T, KANEKO T, NODA K, ITOH T. Augmentation of macrophage functions by an extracellular phosphopolysaccharide from Lactobacillus Delbrueckii ssp. Bulgaricus[J]. Food Microbiology, 2000, 17(1): 109-118. |
| [101] |
刘昭曦, 王禄山, 陈敏. 肠道菌群多糖利用及代谢[J]. 微生物学报, 2021, 61(7): 1816-1828.
LIU ZX, WANG LS, CHEN M. Glycan utilization and metabolism by gut microbiota[J]. Acta Microbiologica Sinica, 2021, 61(7): 1816-1828 (in Chinese). |
| [102] | LI K, LIU XY, ZHANG XL, LIU ZX, YU Y, ZHAO JY, WANG LS, KONG Y, CHEN M. Identification microbial glycans substructure associate with disease and species[J]. Carbohydrate Polymers, 2021, 273: 118595. |
 2023, Vol. 63
2023, Vol. 63